Gluconobacter morbifer G707
A Microbial Biorealm page on the genus Gluconobacter morbifer G707
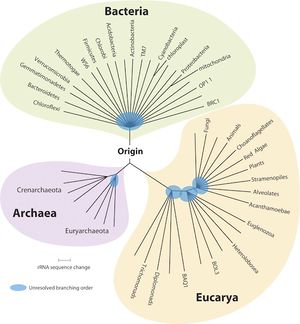
Classification
Higher order taxa
Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconobacter [1]
Species
Gluconobacter morbifer G707 [1]
Description and significance
Gluconobacter morbifer G707 is a non-motile, aerobic, rod-shaped bacteria found in Seoul, South Korea [2]. Isolated from the gastrointestinal tract of the fruit fly Drosophila melanogaster, this bacteria is described as being a commensal symbiont [2]; however, research on the fruit fly’s gut commensal community provides evidence that an abundance of G. morbifer causes apoptosis of gut cells and an early death to its host [3]. In immune-deregulated hosts, G. morbifer is associated with the unique trait of pathogenesis promotion, making this bacteria’s genome sequence particularly valuable in furthering scientific understanding of bacterial pathogenesis in varying host environs [4].
Genome structure
The G. morbifer G707 genome contains 2,899 genes, 50 of which are RNA genes [2]. G+C genome content is 59.04% over 19 DNA scaffolds, all of which are linear [2].
Cell and colony structure
G. morbifer are nonsporulating, rod-shaped bacteria measuring 0.7 by 1.8 to 2.0 um, with circular, pink colonies measuring 1.0 to 1.5 mm in diameter (at 25 degree C and aerobic conditions maintained over the course of a two day period on mannitol-agar) [3]. Some cells stain positive while others stain negative, making this a variable Gram staining bacteria [3]. 3.6 to 7.8 is the recognized optimal pH range for growth [3].
Metabolism
G. morbifer exhibits aerobic respiration using an Ubiquinol oxidation pathway with oxygen and proton transport [2]. G. morbifer is generally auxotrophic, meaning it is unable to manifest many of its own nutritional requirements—like metabolits and amino acids—for growth; however, it is prototrophic for L-aspartate and L-glutamate [2]. This bacteria also is characterized as having an incomplete Coenzyme A biosynthesis [2].
Ecology
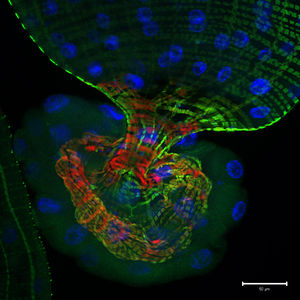
G707, originally isolated in Seoul, South Korea, exhibit endosymbiotic extracellular physical interactions, living commensally within the intestinal [[File:Fruit_Fly_Gut.jpeg|right|300px|A photomicrographic optical section through the tip of a D. melanogaster larval gut. The green shows activity of the Notch signaling pathway; the blue and red represent stained nuclear and cytoskeletal markers respectively. The image was captured by Jessica Von Stetina of Whitehead Institute for Biomedical Research in Cambridge, Massachusetts, USA, and shared by www.nikonsmallworld.com.] microflora of the fruit fly Drosophila melanogaster [2]. Despite being described as commensal, the symbiotic relationship between G. morbifer and
host is ultimately dependent upon the condition of the host’s immunity; the strain will display dominance when the host’s immune homeostasis is compromised, resulting in pathology and gradual host opportunistic infection [4].
Pathology
When inhabiting a Drosophila melanogaster host, mesophile G. morbifer is normally harmless, living commensally with other Drosophila intestinal microbiota [4]. However, when the host’s gut immunity experiences overactivation, G. morbifer becomes dominant, causing apoptosis (programmed cell death) and early termination of the host [4]. With G707’s unique characteristic of becoming pathogenic in an immune-compromised host, its genome sequence possesses potentially substantial value in terms of improving scientific understanding of pathogenesis in varying host environs at the bacterial level [4]. While official results have yet to be published, it is assumed that in further exploration of G. morbifer’s role in fruit fly ecology that a relationship between D. melanogaster physiology and its genotype-based commensal gut community of bacterial strains—specifically with G. morbifer—will be confirmed [3].
References
[1] http://www.ncbi.nlm.nih.gov/bioproject/73361; NCBI
[2] http://img.jgi.doe.gov/cgi-bin/w/main.cgi?section=TaxonDetail&page=taxon Detail&taxon_oid=2513237393; Markowitz1, V., Chen, I., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., Ratner, A., Jacob, B., Huang, J., Williams, P., Huntermann, M., Anderson, I., Mavromatis, K., Ivanova, N., Kyrpides, N. 2012. IMG: the integrated microbial genomes database and comparative analysis system. [Accessed 22 January 2013].
[3] Roh, S., Nam, Y., Chang, H., Kim, K., Kim, M., Ryu, J., Kim, S., Lee, W., Bee, J. 2008. Phylogenic Characterization of Two Novel Commensal Bacteria Involved with Innate Immune Homeostasis in Drosophila melanogaster. Appl Environ Microbio. 74(20): 6171-6177.
[4] Kim, E., Kim, S., Nam, H., Myoung, K., Lee. K., Choi, S., Seo, Y., You, H., Kim, B., Lee, W. 2012. Draft Genome Sequence of Gluconobacter morbifer G707, a Pathogenic Gut Bacterium Isolated from Drosophila melanogaster Intestine. J Bacteriol. 194(5): 1245. Doi: 10.1128/JB.06670-11.
Edited by Lon R. Cameron, student of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio