Hantaviruses: Old World vs. New World
Code Notes:
By Raphael A. Melo
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki.
The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Magnified 20,000X, this colorized scanning electron micrograph (SEM) depicts a grouping of methicillin resistant Staphylococcus aureus (MRSA) bacteria. Photo credit: CDC. Every image requires a link to the source.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Sample citations: [1]
[2]
A citation code consists of a hyperlinked reference within "ref" begin and end codes.
To repeat the citation for other statements, the reference needs to have a names: "<ref name=aa>"
The repeated citation works like this, with a forward slash.[1]
Hantaviruses
Background
Hantaviruses belong to the viral family Bunyaviradae, of which there are five genera: bunyavirus, phlebovirus, nairovirus, tospovirus, and hantavirus (HTV). Although there is no definitive number, there are around 35 known species of HTV that currently exist in the world, including both old-world and new-world strains. Hantaviruses are typically able to infect humans via urine and fecal matter of rodents (Clement et al., 2006). When bedding covered in rodent waste is disturbed, the waste particles become aerosolized and are able to be inhaled, potentially leading to infection (Clement et al., 2006). Rodents are the main vector of this disease into humans, and those who work with rodents that are infected are at an increased risk of becoming infected themselves. Transmission from human to human is far more rare than rodent to human, although it is not impossible. Infected aerosolized particles may also enter the human body through mucous membranes in the eyes, nose, and mouth, and through breaks in the skin, for example, from a rodent bite. Most infections and outbreaks arise within conditions that allow for these particles to become aerosolized, such as during land development, agriculture, and urbanization (CDC, 2021). The first recorded strain was documented as the Hantaan strain, an Old-World variant, between 1951 and 1953 in Korea, and was the first species of many to be documented since. Infecting more than 3000 Korean soldiers during the Korean war, the Hantaan virus outbreak was located near the Hantan river, providing the virus with its name that would later represent all future variants (Mir, 2010). In regards to healthcare, there is no effective cure or treatment for HTV (CDC). Patients will typically receive a broad range of antibiotic therapies and supportive care while being closely monitored (CDC). In regards to treatment based on the type of HTV, there are generally different procedures and drugs administered based on whether or not the species of HTV is “Old World” or “New World.” HTV human infections may result in two possible diseases: hemorrhagic fever with renal syndrome (HFRS) and HPS hantavirus pulmonary syndrome (Avsic-Zupanc et al., 2019). What determines whether the infection yields HFRS or HPS is the type of capillary bed that is infected. For HFRS, renal medulla capillaries are the origin of infection while for HPS, the pulmonary capillaries are the target (Avsic-Zupanc et. al., 2019). Despite this, most initial symptoms tend to be the same for both syndromes include typical flu-like symptoms in addition to aggressive fevers, malaise, and myalgia. Additional symptoms include increased vascular permeability which can lead to hypotension thrombocytopenia and leukocytosis with a left shift, meaning that an abundance of neutrophils are destroyed at the site of infection, causing an increase in production in the bone marrow (Avsic-Zupanc et. al., 2019)(Ishimine et al., 2013).
Genome
Hantaviruses are negative-sensed, enveloped, single-stranded RNA viruses. They have 3 segments of RNA within their genome, Small (S) , Medium (M), and Large (L). Each mRNA segment’s open reading frame is placed in between much smaller non-coding regions that are capped. The L segment plays a key protein in viral replication and codes for the production of L proteins, which facilitate transcriptase and replicase functions. Additionally, L proteins are thought to have a role in endonuclease cleavage of cellular mRNAs to produce primer caps used for transcription initiation of viral mRNAs in a process known as “cap snatching” (CDC, Hantaviruses). The M segment codes for the glycoprotein precursors Gn and Gc, also known as G1 and G2 proteins respectively. These proteins later form the glycoprotein outer membrane matrix and allow the virus to bind to host cell receptors. The S segment codes for N proteins, which serve a variety of functions needed to complete the viral life cycle. Arguably the most important of the proteins, N proteins aid in translation, trafficking, assembly, the possible modulation of the host’s immune system, and serve as the building material needed to form the viral nucleocapsid (Jonsson et al., 2010).
Viral Life Cycle and Structure
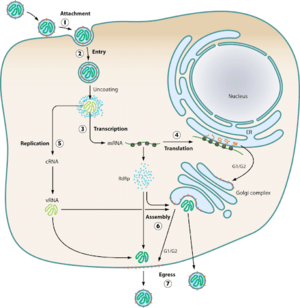
HTV virions typically infect endothelial, epithelial, macrophage, follicular dendritic, and lymphocyte cells (Jonsson et al., 2010). Using glycoproteins, the virions attach to the host cell’s surface membrane receptors and enter the cell via endocytosis. The most common receptors that allow for entry of the virion are β1 integrins, with some exceptions involving β3 receptors (Jonsson et al., 2010). Upon cellular uptake, the virions are enclosed in endosomes where the viral capsule is immediately uncoated. The L, M, and S mRNA segments are set up for transcription via the viral RdSp enzyme, and subsequent translation of the L and S mRNA occurs on free floating ribosomes, while the M mRNA undergoes translation on membrane-bound ribosomes (Jonssonet al., 2010). Assembly of all virion components at the golgi apparatus means the virus exclusively replicates within the host’s cytoplasm (Parvate, 2019). Beyond this, little more than speculation exists on how HTV RNPs are synthesized, trafficked to the golgi, and what drives the budding mechanisms of the golgi. Further speculation of alternative assembly methods for new world hantaviruses could point towards the plasma membrane as a potential assembly site (Jonsson et al., 2010). Golgi vesicles containing newly made viral particles then fuse with the host cell’s plasma membrane, and are then released into the extracellular environment.
Section 2
Include some current research, with at least one figure showing data.
Section 3
Include some current research, with at least one figure showing data.
Section 4
Conclusion
References
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024
