Helicobacter pullorum
NEUF2011
Classification
Higher order taxa
- Domain: Bacteria
- Phylum: Proteobacteria
- Class: Epsilonproteobacteria
- Order: Campylobacterales
- Family: Helicobacteraceae
- Order: Campylobacterales
- Class: Epsilonproteobacteria
- Phylum: Proteobacteria
Species
- Genus: Helicobacter
- Species: pullorum
Helicobacter pullorum; H. pullorum
Description and significance

Helicobacter pullorum is a gram-negative bacteria in the genus Helicobacter. The cells are bacilli with a slight curve. They are typically 3-4 micrometers in length, and 0.3-0.5 micrometers in width. This bacteria species was discovered from chickens, seen in Figure 2, specifically broiler and laying types, and has also been found in humans diagnosed with gastrointestinal disease. H. pullorum thrive in an environment of 37-42 °C and are therefore classified as mesophiles, in the higher end of that temperature range. They grow in microaerobic conditions and are non-spore forming. H. pullorum can also be classified by its response to certain tests. The species is negative for indoxyl acetate esterase, urease and alkaline phophatase production. It tests positive, however, for catalase production. [1, [5]]
They live in the intestinal tract of their host species but can also infiltrate the liver, ultimately causing hepatitis. The species is of significance as it is associated with enteritis and vibrionic hepatitis in poultry. It is also associated with gastroenteritis, diarrhea, liver, and gall bladder disease in humans, and it is speculated that it may play a role in Crohn’s disease. [8]
The amount of infections caused by H. pullorum is most likely underestimated due to its similarities between the genera Helicobacter and Campylobacter. There is a potential for food-borne transmission of H. pullorum to humans, as it has been recognized that Campylobacter species is capable of this. A number of cases concerning the species as a major-food associated human pathogen have been reported, however, there is lack of evidence supporting the prevalence of H. pullorum in humans. [8]
Genome structure
According to a whole genome shotgun sequencing project on Helicobacter pullorum, attained by NCBI, H. pullorum has a circular genome consisting of 1,919,070 nucleotides with a known sequence. The DNA codes for 2044 genes, 2008 of which are protein coding and 36 of which are structural RNAs. 33% of the genome consists of GC pairings. [12]
Cell structure and metabolism
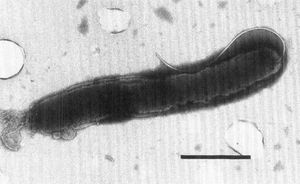
Being Gram negative, H. pullorum has two cell membranes. The cell is unable to grow at 25 °C, but grows at 37 °C under microaerobic conditions. Under aerobic conditions at 37 °C, H. pullorum will not grow. Most Helicobacter cells have bundles of multiple sheathed flagella. However, H. pullorum has one single flagellum that is nonsheathed, as seen in Figure 1. H. pullorum produces catalase, reduces nitrates, and does not produce urease or alkaline phosphatase. It also produces cytolethal distending toxin, which causes distension in certain cell lines, causing the cell to disintegrate and die. [6]
Ecology
Mucous is the natural niche of the genus helicobacter. The prevalence of colonized spiral shaped bacteria in the gastric mucosa of most animals demonstrates this. It is likely that the spiral shape of the bacteria give it a selective advantage in this viscous environment. H. pullorum is a member of the enterohepatic helicobacters, which colonizes mainly in the intestine and the hepatobiliary system. The species is naturally found in chickens and birds, and was originally isolated from chickens. There is a possibility that humans may have helicobacter as a part of their normal flora in their lower bowel. [4] However, there is lack of evidence to support this idea. H. pullorum has also been isolated in fecal samples from humans with gastroenteritis, as well as from the gallbladders of women who had chronic cholecystitis, and from the livers of patients suffering from cirrhosis or hepatocellular carcinoma. [3]
Pathology
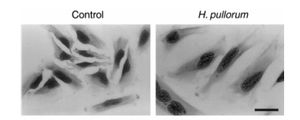
Very little is known about the pathology of H. pullorum. There is a lack of evidence as to whether the bacteria is harmful to its host. It is known to grow primarily in the intestinal tract of chickens, but has also been found in the feces of humans. A study conducted in 2005 showed that H. pullorum was present in roughly the same percentage of fecal samples from patients with gastrointestinal disease (4.3%) as there were in healthy patients (4.0%). This does not necessarily mean that H. pullorum is not a potential cause of gastrointestinal disease. It could be that, like many bacteria, there are strains of H. pullorum that are virulent, while others are not. This is supported by a study conducted in 2000 that showed that H. pullorum can produce cytolethal distending toxin, which is a potential virulence factor that may or may not be a direct cause of diarrhea. The effects of cytolethal distending toxin, including cytoplasmic distension and nuclear enlargement, are seen in Figure 3. [2][8]
Current Research
Helicobacter pullorum is a relatively new species (discovered in 1994), and because of this much of the research done on this organism has involved its prevalence in turkeys. A study done by Zanoni, Piva, Rossi & Pasquali (2011), experimented with H. pullorum and its antibiotic resistance. Researchers used 55 turkeys from 11 farms in northern Italy for this experiment. Colonies were identified by PCR assay before being tested for resistance. Each of the 11 farms had animals positive for H. pullorum with a total of 42 out of the 55 animals infected. All isolates tested were susceptible to gentamycin and chloramphenicol. The majority of resistance was observed with erythromycin (9 isolates), nalidixic acid (8 isolates), and ciprofloxacin (8 isolates), with multi-resistance seen between erythromycin and ciprofloxacin antibiotics. In addition, a high prevalence of horizontal gene transfer and recombination was observed in the species. The combination of high frequency of isolation with multiple antibiotic resistance suggests that H. pullorum could be a major risk factor for contamination if found to be transmitted through food. Further testing needs to be done to determine if H. pullorum can be spread as a food-born illness, and if it has antibiotic resistance in human hosts. [9]
Helicobacter pullorum has been isolated from the ceca of poultry, livers of hens with lesions suggestive of hepatitis, and the feces of human patients with gastroenteritis. H. pullorum shares characteristics with Campylobacter species, making the two commonly confused. It is thought, therefore, that the prevalence of H. pullorum is underrepresented. Cytolethal distending toxin (CDT) has been found in several Helicobacter bacteria. The activity of CDT causes appearance of cellular distension, irregularities in the cytoskeleton, and lethality. The study done by Young, et al determined whether CDT activity was present in H. pullorum. The gene, cdtB, was found. H. pullorum produced cellular distension and cell cycle arrest, identical to bacteria that are known to have CDT activity. H. pullorum samples were also taken from humans with gastroenteritis, as clinical examples, and all were positive for the presence of the gene for CDT expression. The presence of this gene is thought to be a potential marker for identifying H. pullorum, along with using already known phenotypic and genotypic traits. [8]
Helicobacter pullorum has been linked with inflammatory bowel disease (IBD), but there is no evidence to prove that the bacteria is a potential cause. Previous research has implied that H. pullorum may have pro-inflammatory properties on epithelial cells. Although the H. pullorum genome has not yet been sequenced, some virulence markers have been identified, suggesting that the microorganism may be pathogenic. Avian and human H. pullorum strains can activate translocation of the nuclear factor-kB (NF-kB), which is an essential transcription factor in chronic inflammatory disease. NF-kB, once inside the cytoplasm of a cell, can move into the nucleus and regulate the expression of multiple target genes, such as interleukin 8 (IL8). A study done by Varon, et al aimed to establish whether H. pullorum is capable of having a direct effect on human intestinal epithelial cells in vitro. The study also sought to determine the bacterial mechanisms and the signaling pathway involved in H. pullorum. To accomplish this, the study measured the pro-inflammatory properties of the microorganism on human gastric and intestinal epithelial cell lines. The results showed that all of the H. pullorum strains that were tested stimulated IL8 secretion, heat-killed H.pullorumhad the same effect as well. Incubation of cells that were filtered of H.pullorum culture supernatants did not stimulate IL8 secretion. The experiment also demonstrated that the bacteria induced NF-kB activation as well as nuclear translocation, which was seen by immunofluorescent staining and cellular fractionation. The experiment concluded that H. pullorum strains are mediated by NF-kB signaling and stimulates IL8 secretion by way of human gastric and intestinal epithelial cell lines. This research supports the case that H. pullorum is a human pathogen, and has a role in acute and chronic digestive disease. [7]
Researchers have developed a method for real-time, direct detection of Helicobacter pullorum (and close relatives) in food using a new PCR-assay. Using this real-time assay, the authors of the experiment then tested for the presence of H. pullorum in food samples from Valencia. Using both the real-time assay and the “regular” PCR assay on all thirty chicken samples of varying types (livers, carcasses, chicken burgers), there were found to be no culturable samples of H. pullorum in the food. However, 37% of the total samples were found to have “H. pullorum-like DNA” in them. Upon further analysis, it was determined that the DNA was that of H. pullorum due to the 99% concordance to the 16S ribosomal DNA gene. As detection of H. pullorum is still inconsistent at this point, the idea is that the novel real-time PCR assay will hopefully allow a larger range of detection so the bacteria can be studied more in-depth. [11]
Cool Factor
Helicobacter pullorum demonstrates cytotoxic activity causing cellular distention and halting of mitosis at G2/M stage. It is thought that this cytotoxic activity is what gives bacteria such as H. pullorum their pathogenicity in eukaryotic cells such as those of a chicken and a human. The cytotoxic activity of H. pullorum is thought to target lymphocytes, causing a decreased immune response in the body and ultimately sickness. [1]
References
[10] http://envirowiki.unitycollege.wikispaces.net/Chickens
[12] http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=6018
