Herminiimonas glaciei: Difference between revisions
No edit summary |
|||
| Line 4: | Line 4: | ||
==Higher order taxa== | ==Higher order taxa== | ||
Domain: Bacteria; Phylum: Proteobacteria; Class: Betaproteobacteria; Order: Burkholderiales; Family: Oxalobacteraceae; Genus: Herminiimonas. | Domain: Bacteria; Phylum: Proteobacteria; Class: Betaproteobacteria; Order: Burkholderiales; Family: Oxalobacteraceae; Genus: Herminiimonas. | ||
==Species== | ==Species== | ||
Herminiimonas glaciei (strain UMB49). | Herminiimonas glaciei (strain UMB49). | ||
[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?&id=523788 NCBI Taxonomy ID: 523788] | [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?&id=523788 NCBI Taxonomy ID: 523788]. | ||
=Description and Significance= | =Description and Significance= | ||
Revision as of 03:29, 15 December 2012
Classification
Higher order taxa
Domain: Bacteria; Phylum: Proteobacteria; Class: Betaproteobacteria; Order: Burkholderiales; Family: Oxalobacteraceae; Genus: Herminiimonas.
Species
Herminiimonas glaciei (strain UMB49).
Description and Significance
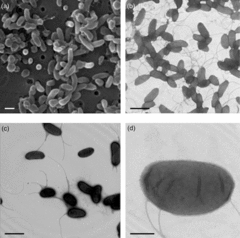
Herminiimonas glaciei (as strain UMB49) is a gram-negative ultramicrobacterium isolated from 120,000-year old ice ~3km within a Greenland glacier [1]. Originally recovered in a dormant form, the bacterium was revived after long-term enrichment, involving a 7 month incubation at 2oC in anaerobic conditions, and nearly 5 month of incubation on agar plates at 5oC [2]. This strain has been classified as a unique species within the recently formed genus Herminiimonas [2] and is most closely related to H. saxobsidens [3], one of the four other recognized species in the genus (the rest are H. arsenicoxydans, H. fonticola and H. aquatilis [2]).
Bacteria such as H. glaciei that can remain alive in ice for extended periods of time are of great significance to Astrobiology [5], since extremely cold environments are the best analogues of possible extraterrestrial habitats [4]. Loveland-Curtze, who headed the team that discovered the species, speculates that the study of H. glaciei and other ultramicrobacteria and extremophiles may offer insight into the existence of microorganisms on other planets [6], by elucidating the cell mechanisms used to survive extremely harsh conditions [4]. An understanding of the nature and evolution of ultramicrobacteria like H. glaciei is also essential for the resolution of important biological questions, including the concept and mechanisms of microbial cell minimization, the minimum permissible sizes of living cells, and the origin of living organisms [7].
Genome Structure
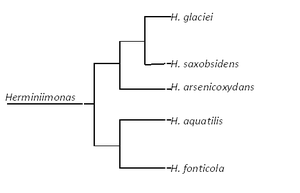
The complete genome of H. glaciei has not yet been sequenced. However, phylogenetic analyses of 16S rRNA gene sequences have shown a high degree of relatedness between H. glaciei (strain UMB49) and the four other species of Herminiimonas (97.6-99.6%) [2]. In contrast, DNA-DNA hybridization experiments between genomic DNA from H. glaciei and its closest relatives implied low DNA-DNA relatedness (45-57%). Contrasting high 16S rRNA gene sequence similarity and low DNA-DNA relatedness were also reported for other Herminiimonas species [10].
H. glaciei’s genome size likely falls between 0.58-3.2Mb, given that the average cell has a volume of 0.043μm3 – Since it classifies as an ultramicrobacterium based on the cell volume criterion (<0.1μm3) [8], it is expected that the bacterium’s genome size falls within the specified range (3.2-0.58Mb) [9] for ultramicrobacteria as well. H. glaciei’s DNA G+C content is 59.0mol%.
Cell Structure, Metabolism and Life Cycle

H. glaciei is a small non-sporulating rod with several long flagella extending from the sides and ends of the cell. These structures visually differentiate H. glaciei from the four other Herminiimonas species, and aid in the swarming motility displayed by the bacteria [2]. With cellular dimensions of 0.5-0.9 by 0.3-0.4μm H. glaciei is roughly 10-50 times smaller than E. coli [2], making it small even by bacterial standards. The miniscule size of H. glaciei is probably what permits it to survive in small, briny, liquid veins between ice crystals [4]. The original colonies grown from the ice were small and purplish-brown, but aerobic recultivation resulted in translucent colonies ranging between white to tan in colour.
H. glaciei is capable of growth both aerobically and microaerophilically, whereas the rest of the recognized Herminiimonas species are aerobes [2]. H. glaciei also doesn’t reduce nitrate nor does it use formate, although type strains of the other Herminiimonas species do [3]. H. glaciei does degrade fatty acids however, and its fatty acid profile is similar to those of its relatives. It also grows in minimalistic salts media in the absence of vitamin supplementations, like most of its recognized relatives [11] [12] which grow slowly if at all on rich media.
The growth rate of H. glaciei peaks at a temperature of 30oC, with the bacteria doubling every 4 hours at that temperature. Growth occurs anywhere from 1oC-35oC however, making H. glaciei a psychrotolerant mesophile [2]. Not much is known about the life cycle of H. glaciei to date.
Ecology

The only known habitat of H. glaciei is the 3000m-deep layer of glacial ice from which it was isolated in Greenland. It survived in a dormant form in tiny liquid veins in the ice for 120,000 years, a feat made possible by the bacterium’s miniscule dimensions [4]. Small cell size is also advantageous for more efficient nutrient uptake and micro-niche occupation [7], and probably contributed to the bacterium’s ability to sustain itself in a very nutrient-poor environment. An important point to keep in mind is that though H. glaciei was able to survive glacial temperatures, when revived it was found to grow optimally at 30oC. This suggests that its original environment may have been drastically different from the ice in which it was found – in fact, its closest relative H. saxobsidens was isolated from a lichen-covered rock [3]. In any case, H. glaciei`s ability to remain alive in the ice for such a long time may have important implications about the possibility of life in the extreme cold of extraterrestrial habitats,

for example in the Mars polar ice caps or in the icy oceans of Jupiter’s moon Europa [6].
Interestingly, despite having been frozen 3km underground for 120,000 years, H. glaciei is resistant to a number of modern antibiotics including ampicillin, ciprofloxacin, penicillin, and vancomycin [2]. The bacterium can also pass through 0.2-micron filters, which are the standard filter pore size used for the sterilization of fluids in laboratories and hospitals. Though H. glaciei is not pathogenic, Loveland-Curtze speculates that if there are pathogenic ultramicrobacteria, their potential presence in solutions presumed to be sterile would pose a huge problem as the tiny cells would be able to replicate and still not have sufficient density to turn a clear solution cloudy [4].
References
(1) Miteva, V.I. and Brenchley, J.E. (2005). “Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core”. Applied and Environmental Microbiology, 71(12): 7806-18. doi:10.1128/AEM.71.12.7806-7818.2005
(2) Loveland-Curtze, J., Miteva, V.I. and Brenchley, J.E. (2009). “Herminiimonas glaciei sp. nov., a novel ultramicrobacterium from 3042 m deep Greenland glacial ice”. International Journal of Systematic and Evolutionary Microbiology, 59(6): 1272-7. doi:10.1099/ijs.0.001685-0
(3) Lang, E., Swiderski, J., Stackebrandt, E., Schumann, P., Spröer, C. and Sahin, N. (2007). “Herminiimonas saxobsidens sp. nov., isolated from a lichen-colonized rock”. International Journal of Systematic and Evolutionary Microbiology, 57(11): 2618-22. doi:10.1099/ijs.0.65163-0
(4) (2009). “Herminiimonas Glaciei - Tiny frozen microbe may be clue to extra-terrestrial life”. Science 2.0. Retrieved 2012-10-26.
(5) Hoover, R.B. and Pikuta, E.V. (2009). “Life in Ice: Implications to Astrobiology”. Instruments and Methods for Astrobiology and Planetary Missions, XII Proc. SPIE7441. doi:10.1117/12.832640
(6) Colgan, A. (2009). “‘Resurrection bug’ revived after 120,000 years”. New Scientist. Retrieved 2012-10-26.
(7) Duda, V.I., Suzina, N.E., Polivtseva, V.N. and Boronin, A.M. (2012). “Ultramicrobacteria: Formation of the Concept and Contribution of Ultramicrobacteria to Biology”. Microbiology, 81(4): 379-90. doi:10.1134/S0026261712040054
(8) MacDonell, M.T. and Hood, M.A. (1982). “Isolation and characterization of ultramicrobacteria from a gulf estuary”. Applied and Environmental Microbiology, 43(3): 556-571. PMCID: PMC241875
(9) Velimirov, B. (2001). “Nanobacteria, Ultramicrobacteria, and Starvation Forms: A search for the smallest metabolizing bacterium”. Microbes and Environments, 16(2): 67-77. doi:10.1264/jsme2.2001.67
(10) Kämpfer, P., Busse, H-J. and Falsen, E. (2006). “Herminiimonas aquatillis sp. nov., a new species from drinking water”. Systematic and Applied Microbiology, 29,(4): 287-91. PMID:16436326
(11) Muller, D., Simeonova, D.D., Riegel, P., Mangenot, S., Koechler, S., Lièvremont, D., Bertin, P.N. and Lett, M-C. (2006). “Herminiimonas arsenicoxydans sp. nov., a metalloresistant bacterium”. International Journal of Systematic and Evolutionary Microbiology, 56(8): 1765-9. doi:10.1099/ijs.0.64308-0
(12) Fernandes, C., Rainey, F.A., Nobre, M.F., Pinhal, I., Folhas, F. and da Costa, M.S. (2005). “Herminiimonas fonticola gen. nov., sp. nov., a Betaproteobacterium isolated from a source of bottled mineral water. Systematic and Applied Microbiology, 28(7): 596-603. PMID:16156117
