Hospital-acquired Methicillin Resistant Staphylococcus Aureus (MRSA): Difference between revisions
Alexandera (talk | contribs) |
Alexandera (talk | contribs) |
||
| Line 13: | Line 13: | ||
<br>High replication rates coupled with the great ability of to perform horizontal gene transfer (especially through conjugation) allow bacteria to develop antibiotic resistance and to spread it quickly. By 1942, the first penicillin resistant strains of <i>Staphylococcus aureus</i> had been isolated in hospitals (4). Theses penicillin resistant strands contained a plasmid encoding a penicillin-hydrolyzing enzyme, penicillinase. Less than 20 years after the first strains of <i>Staphylococcus aureus</i> were found to be resistant to penicillin, 80% of all strains had acquired penicillin resistance. As new antibiotics such as methicillin and vancomycin were used to fight <i>Staphylococcus aureus</i>, resistance to these antibiotics also began to develop. Methicillin was first used to treat <i>Staphylococcus aureus</i> in 1959 and just after 2 years of use, methiccillin-resistant <i>Staphylococcus aureus</i> (MRSA) strains had be isolated (4). Methicillin resistance first developed and became transferable in the mecA gene. The mecA gene encodes a protein, penicillin-binding protein PBP2a, which cannot be bound by Beta-lactam antibiotics (penicillin, methicillin…) and in turn prevents the disruption of cell wall formation by these antibiotics. This gene is located on mobile genetic element called the Staphylococcal Cassette Chromosome mec (SCCmec)(4). | <br>High replication rates coupled with the great ability of to perform horizontal gene transfer (especially through conjugation) allow bacteria to develop antibiotic resistance and to spread it quickly. By 1942, the first penicillin resistant strains of <i>Staphylococcus aureus</i> had been isolated in hospitals (4). Theses penicillin resistant strands contained a plasmid encoding a penicillin-hydrolyzing enzyme, penicillinase. Less than 20 years after the first strains of <i>Staphylococcus aureus</i> were found to be resistant to penicillin, 80% of all strains had acquired penicillin resistance. As new antibiotics such as methicillin and vancomycin were used to fight <i>Staphylococcus aureus</i>, resistance to these antibiotics also began to develop. Methicillin was first used to treat <i>Staphylococcus aureus</i> in 1959 and just after 2 years of use, methiccillin-resistant <i>Staphylococcus aureus</i> (MRSA) strains had be isolated (4). Methicillin resistance first developed and became transferable in the mecA gene. The mecA gene encodes a protein, penicillin-binding protein PBP2a, which cannot be bound by Beta-lactam antibiotics (penicillin, methicillin…) and in turn prevents the disruption of cell wall formation by these antibiotics. This gene is located on mobile genetic element called the Staphylococcal Cassette Chromosome mec (SCCmec)(4). | ||
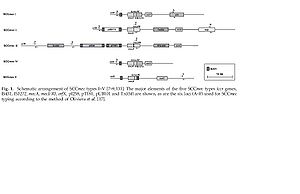
[[Image:5_types_of_sccmec.jpg|thumb|800x517|right|Schematic arrangement of the 5 types of SCCmec providing methicillin resistance in <i>Staphylococcus aureus</i>]] | <br>[[Image:5_types_of_sccmec.jpg|thumb|800x517|right|Schematic arrangement of the 5 types of SCCmec providing methicillin resistance in <i>Staphylococcus aureus</i>]]<br> | ||
Five types (Types I-V) of SCCmec have been isolated and characterized. They vary in size from 20.9 to 66.9 kb. The five SCCmec types were isolated sequentially as one would expect, but they were each isolated initially in different regions across the globe. SCCmec Type I was isolated in 1961 in the UK, Type II in 1982 in Japan, Type III in 1985 in New Zealand and finally Type V at the start of the 21st century in Australia (4). Not only might one find it surprising that the different types of SCCmec would have been isolated in areas so far apart from each other, but it is also interesting that not all of the different SCCmecs encode/provide the same resistance. Types I, IV and V only encode resistance to beta-lactam antibiotics where as Types II and III encode additional drug resistance genes. The additional drug resistance genes include the transposon Tn554 which codes for an ermA gene providing inducible resistance to macrolide, lincosamide and streptogramin(4). So how did the first MRSA isolates lead to the development of the MRSA isolates found today? | Five types (Types I-V) of SCCmec have been isolated and characterized. They vary in size from 20.9 to 66.9 kb. The five SCCmec types were isolated sequentially as one would expect, but they were each isolated initially in different regions across the globe. SCCmec Type I was isolated in 1961 in the UK, Type II in 1982 in Japan, Type III in 1985 in New Zealand and finally Type V at the start of the 21st century in Australia (4). Not only might one find it surprising that the different types of SCCmec would have been isolated in areas so far apart from each other, but it is also interesting that not all of the different SCCmecs encode/provide the same resistance. Types I, IV and V only encode resistance to beta-lactam antibiotics where as Types II and III encode additional drug resistance genes. The additional drug resistance genes include the transposon Tn554 which codes for an ermA gene providing inducible resistance to macrolide, lincosamide and streptogramin(4). So how did the first MRSA isolates lead to the development of the MRSA isolates found today? | ||
Revision as of 02:08, 16 April 2009
By: Anthony Alexander
Introduction
Staphylococcus aureus is a spherical microbe and a member of the bacteria domain. This bacterium can be found naturally on the skin and in the mucus membranes of humans most importantly. In fact, Staphylococcus aureus can be found in the nostrils of up to 30% of people (1). The bacteria is spread most commonly through human contact be it hand-to-hand, from a wound secretion or mucus.
The spherical bacteria is gram-positive (contains a peptidoglycan layer in its cell wall) and forms colonies that grow in two planes (2). Staphylococcus aureus however lacks flagella and as a result is non-motile. The genome of Staphylococcus aureus was first sequenced and mapped by Peter A. Pattee for the strain NCTC 8325 (2). This strain had a circular genome containing approximately 2,900 open reading frames, 61 tRNA genes, 3 structural RNAs and 5 complete ribosomal RNA operons (2). The aspect of Staphylococcus aureus and its genome that is most concerning revolves around the plasmids that are incorporated/associated with this bacterium’s genome. The plasmids that this bacterium contains often code some type of antibiotic resistance. Over time, Staphylococcus aureus was able to acquire antibiotic resistance through conjugation (horizontal gene transfer) of a plasmid containing a transposon, Tn 1546 (2). The transposon Tn 1546 contains the gene mecA which encodes a modified penicillin-binding protein granting Staphylococcus aureus methicillin, and more broadly, penicillin resistance.
Methicillin is a beta-lactam antibiotic similar to penicillin. Beta-lactam antibiotics target penicillin-binding proteins. Once bound to the protein, the antibiotic prevents proper peptidoglycan and cell wall formation so that cells will eventually burst as the bacteria attempt to grow larger (3). Penicillin and similar beta-lactam ring antibiotic resistance can be achieved in two main ways; first bacteria can acquire a gene (through horizontal gene transfer) encoding beta-lactamase enzyme which cleaves the critical ring structure of this particular type of antibiotics preventing them from binding the penicillin-binding proteins and disrupting cell wall formation. Second, some bacteria can produce a modified penicillin-binding protein that no longer actually binds the antibiotic which again prevents the desired effects of the antibiotic (3). Methicillin is frequently used to fight bacteria that produce beta-lactamase but is still susceptible to the second form of bacterial resistance to beta-lactam antibiotics. Bacteria like methicillin resistant Staphylococcus aureus are currently of special interest, especially in hospitals, because very few drugs (antibiotics) are still effective against them.
Antibiotic Resistance in Staphylococcus aureus in Greater Detail
High replication rates coupled with the great ability of to perform horizontal gene transfer (especially through conjugation) allow bacteria to develop antibiotic resistance and to spread it quickly. By 1942, the first penicillin resistant strains of Staphylococcus aureus had been isolated in hospitals (4). Theses penicillin resistant strands contained a plasmid encoding a penicillin-hydrolyzing enzyme, penicillinase. Less than 20 years after the first strains of Staphylococcus aureus were found to be resistant to penicillin, 80% of all strains had acquired penicillin resistance. As new antibiotics such as methicillin and vancomycin were used to fight Staphylococcus aureus, resistance to these antibiotics also began to develop. Methicillin was first used to treat Staphylococcus aureus in 1959 and just after 2 years of use, methiccillin-resistant Staphylococcus aureus (MRSA) strains had be isolated (4). Methicillin resistance first developed and became transferable in the mecA gene. The mecA gene encodes a protein, penicillin-binding protein PBP2a, which cannot be bound by Beta-lactam antibiotics (penicillin, methicillin…) and in turn prevents the disruption of cell wall formation by these antibiotics. This gene is located on mobile genetic element called the Staphylococcal Cassette Chromosome mec (SCCmec)(4).
Five types (Types I-V) of SCCmec have been isolated and characterized. They vary in size from 20.9 to 66.9 kb. The five SCCmec types were isolated sequentially as one would expect, but they were each isolated initially in different regions across the globe. SCCmec Type I was isolated in 1961 in the UK, Type II in 1982 in Japan, Type III in 1985 in New Zealand and finally Type V at the start of the 21st century in Australia (4). Not only might one find it surprising that the different types of SCCmec would have been isolated in areas so far apart from each other, but it is also interesting that not all of the different SCCmecs encode/provide the same resistance. Types I, IV and V only encode resistance to beta-lactam antibiotics where as Types II and III encode additional drug resistance genes. The additional drug resistance genes include the transposon Tn554 which codes for an ermA gene providing inducible resistance to macrolide, lincosamide and streptogramin(4). So how did the first MRSA isolates lead to the development of the MRSA isolates found today?
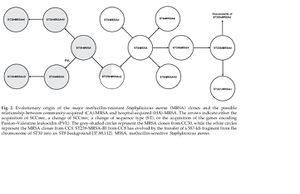
In a paper by Deurenberg et al. two theories establishing the relationship between the first MRSA strains and present day MRSA strains are proposed. The first is called the single-clone theory which states that all MRSA clones or present day strains have a common ancestor. This first theory also states that SCCmec was introduced only once into Staphylococcus aureus. After its “introduction” or formation, SCCmec developed into types I-V through mutations overtime. The second theory is called the multi-clone theory. This second theory suggests that SCCmec was introduced several times into different Staphylococcus aureus. According to the paper by Deurenberg et al. the multi-clone theory has received greater support recently. It appears as though the different SCCmec tpes have been acquired by Staphylococcus aureus strains with different genetic backgrounds. This is also a likely reason as to why the different types were first isolated in regions so far apart from each other.
Prevalence, Type and Spread of MRSA
As described previously, once an antibiotic resistance is developed, it can be transferred and spread through bacteria strains very rapidly. In England and Wales, less than 2% of Staphylococcus aureus strains were methicillin-resistant in 1990 but by 2002 42% of Staphylococcus aureus strains were methicillin-resistant (5). In a paper by Carnicer-Pont et al. it is stated that an estimated 300,000 cases of hospital-acquired MRSA occur each year in England leading to 5,000 deaths. The main focus of the paper by Carnicer-Pont et al. is achieving a better understanding of how MRSA is spread within hospitals. From the study, it was determined for the hospital in which the study took place that MRSA strains were most frequently acquired as a result of surgical procedures. The insertion of urinary catheters and central lines accounted for 51% and 39% of the MRSA acquired cases respectively. Surgical site infection accounted for the lowest percent of the cases (16%) but time at risk accounted for 36% of the cases. In the study time at risk was considered to be a period of being exposed to or in condition allowing the spread of MRSA for a period greater than 7 days. If not appropriately accounted for, MRSA strains can spread easily and quickly in a hospital setting.
The spread of MRSA within hospitals is not the only concern however. In a study done throughout 4 of 34 German university hospitals by Chaberny et al. the type of MRSA infections were studied. The majority of MRSA infections were wound infections (56.9%) with pneumonia cases being the second most common (21.0%). Potentially the most dangerous infection type, bloodstream infections accounted for 15.1% of the cases and urinary track infection accounted for 6.9%. One of the most important pieces of information provided by this study was the fact that of all the infected MRSA patients, 51.5% had already been infected at their time of admission. This suggests that not only should the spread of MRSA infections to non-infected patients in hospitals be a major concern, but that the introduction of new MRSA strains to hospitals by patients infected prior to admission should be a concern as well. So how can hospitals deal with the introduction of new MRSA strains and the spread of MRSA within their buildings? Some of the best ways to prevent the spread of MRSA is through screening and resulting isolation.
Fighting the Spread of MRSA in Hospitals
A study by Mahamat et al. (2007) set out to determine the most effective methods of preventing the spread of MRSA in hospitals. The study used a control hospital (which received a new antibiotic treatment policy as well as an alcohol hand gel policy) and an intervention hospital (which received the antibiotic treatment policy, alcohol hand gel policy as well as environmental screening, chlorine disinfection and admission screening) to determine the relative effectiveness of these policies. The new antibiotic treatment policies did not prove to be an effective way of fighting the spread of MRSA infections in hospitals. The introduction of alcohol hand gel for improved hand hygiene did however prove to be very effective in reducing the spread of MRSA. In the control hospital (alcohol hand gel introduced after new antibiotic treatment policy) there was a 30% decrease in the spread of MRSA in the hospital. In the intervention hospital the introduction of alcohol hand gel reduced the spread of MRSA by 21% (9). The decrease experienced in the intervention hospital was likely smaller than that compared to the control hospital because the prevention measures of environmental swabbing for MRSA as well as chlorine disinfection of environments contributed to a 32% decrease in the spread of MRSA and these measures were not taken in the control hospital(9). The admission screening administered in the intervention hospital was associated with 22% of the decrease in the spread of MRSA in the hospital. As can be seen in the provided figure, combining the spread prevention techniques described above as in the intervention hospital is most effective in preventing the spread of MRSA in hospitals. For this reason, improved hand hygiene and admission screening along with their effects in other studies will be focused on later in this section.
The decision to fight MRSA in hospitals revolves around three basic questions. First, is MRSA that much worse than MSSA? Second, how effective can we be in reducing the spread of MRSA? Lastly, is fighting MRSA cost effective?
Both methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA) can lead to serious infections, food poisoning and even toxic shock syndrome. Although infections by both can be dangerous, as would be expected given their names, MSSA infections are much easier to treat than MRSA infections. Therefore, as one might expect, patients with MRSA bacteraemia had a three-fold increase in cost and a three-fold increase of the length of the hospital stay when compared to those with MSSA infections (7). The length of hospital stay is much longer for MRSA infections than MSSA infections for a number of reasons. Most importantly, the drugs most commonly used to fight MRSA infections, glycopeptides like vancomycin, are much less effective at fighting Staphylococcus aureus infections than beta-lactam antibiotics are against fighting MSSA infections (7). As a result, treatment may not completely resolve MRSA infections allowing them to relapse and in turn extending drug treatment plans as well as extended or even repeated hospitalizations. The repeated use of drugs to fight MRSA infections increases the likelihood that MRSA will also develop resistance to these drugs as well. As stated previously, some Staphylococcus aureus strains are becoming resistant to vancomycin through vanA. Resistance to vancomycin as well as the beta-lactam antibiotics (penicillin, methicillin…) would make fighting Staphylococcus aureus infections even more difficult than fighting MRSA infections. MRSA spread should be fought not only because it takes longer and is harder to fight than MSSA, but MRSA control can also have a positive affect on the rate of other hospital-acquired infections. Studies have shown that control measures used to decrease rates of MRSA also decreased the rates of other hospital-acquired infections due to other pathogens (7).
After concluding that fighting the spread of and treating MRSA properly is crucial, can we be effective in preventing the spread of MRSA in hospitals? A review by Rubinovitch and Pittet (2001) provides a number of studies in which hospitals were able to significantly reduce the spread of MRSA in hospitals. The greatest effects were seen in a study described in the review that took place at the Iowa City Veteran's Affairs Medical Centre. At this medical center, what was described as a simple control plan including the isolation of MRSA infected patients and an increased emphasis on hand-washing and hygiene led to a 50% reduction in the number of hospital-acquired MRSA infections (7). Rapid screening for MRSA in newly admitted patients allows for contact isolation of MRSA strains. When proper hygiene techniques are followed after MRSA strains have been isolated, the spread of these MRSA strains throughout the hospital is extremely reduced. Early detection of MRSA infected patients that are admitted to hospitals was found to decrease the total number of MRSA isolation days by 42% and prevent 8 to 41 hospital-acquired MRSA infections per year in hospitals (7). Clearly early screening for MRSA colonization allows for the rapid detection of MRSA infections allowing for these cases to be isolated and dealt with using emphasized proper hygiene protocols and ultimately limiting the spread of MRSA in hospitals.
Despite knowing that hospitals should prevent the spread of MRSA infections due to the risk of even further drug resistance over MSSA strains, and that hospitals can be effective in preventing the spread of MRSA infections, is it cost-effective for hospitals to implement the screening and isolation processes needed to fight the spread of MRSA even though Staphylococcus aureus infections will undoubtedly occur in hospitals? In other words, is the cost of hosting patients for longer periods of time, the costs associated with isolating patients, and the increased cost of treating MRSA infections more or less than the costs of screening newly admitted patients? If the costs of selective screening combined with the treatment of these MRSA infections is less than the cost of treating MRSA infections as well as MRSA infections that spread throughout the hospital when screening wasn’t done, screening would be considered cost effective. A study in Germany by Wernitz et al. (2005) attempted to determine whether hospital wide selective screening for MRSA colonization would be cost-effective. A selective-screening program was found to be cost-effective at the point where 2.9 cases of hospital-acquired MRSA infections were prevented per year. As was stated previously, early screening/detection of MRSA colonized patients could prevent the spread of 8 to 41 hospital-acquired MRSA infections per year. Combining these statistics suggest that in areas of endemic MRSA infections, a selective-screening program would be extremely cost-effective. In fact, the study by Wernitz et al. (2005) established that the screening of all incoming for MRSA colonization would become cost-effective if at least 22% of the patients screened were MRSA positive. Therefore, the amount of screening or the selectivity of screening done for MRSA colonization depends on the frequency at which hospitals admit or treat MRSA colonized patients.
Synergistic Treatment of MRSA
The continued development of resistance to more and more drugs makes the treatment of Staphylococcus aureus infections and especially MRSA infections is becoming increasingly difficult. Recent studies involving the use of naturally occurring substances to fight MRSA have provided great insight as to what direction the fight against MRSA will be heading in. Although the use of these naturally occurring substances may require high doses to be effective against MRSA when used by themselves, synergistic use with antibiotics such as oxacillin proves to be extremely effective.
A study by Hatano et al. (2005) examines the effects of a number of these naturally occurring substances and flavonoids found to be directly effective against MRSA infections. From their study, the licorice herb emerged as one of the strongest candidates for synergistic use with antibiotics to fight MRSA infections. Licorice produces an isoflavan called licoricidin which is thought to alter the inhibitory effect of PBP2a (peptidoglycan-binding-protein 2a) on cell wall formation. When coupled with oxacillin, licoricidin substantially reduced the minimum inhibitory concentration (MIC) of oxacillin required to effectively combat MRSA infections. A licoricidin concentration of 4 ug/mL reduced the MIC of oxacillin from 128-256 ug/mL to 8-16 ug/mL and a licoricidin concentration of 8 ug/mL reduced the MIC of oxacillin to less than 0.5 ug/mL. Products from the breakdown of tea polyphenols were also found to lower the MIC of oxacillin needed to fight MRSA infections.
In 2008, Cho et al. released a paper/study further examining the use of tea polyphenols (TPP) in synergy with oxacillin and MRSA infections. Green tea polyphenols such as epigallocatechin gallate (EGCG) are noted as having bactericidal activity against gram-positive bacterium such as Staphylococcus aureus. The antibacterial and syngergistic activities of TPP with oxacillin were examined on 13 clinical isolates of MRSA. As a bactericidal, TPP required a MIC ranging from 50-180 ug/mL to fight MRSA alone. When TPP was used synergistically with oxacillin, the required MICs were reduced significantly for all 13 strains of MRSA. A 0.5xMIC of TPP (60ug/mL) added to a 0.25xMIC of oxacillin (64ug/mL) killed almost all of the MRSA bacteria within 24 hrs of incubation. Studies like those done by Cho et al. and Hatano et al. are at the foremost front of research in developing methods of fight Staphylococcus aureus and especially methicillin-resistant Staphylococcus aureus infections.
Conclusions
Staphylococcus aureus is a bacterium that naturally inhabits the skin and nose of humans. If the bacterium is able to enter the body (often through wounds or sores) it can cause a number of infections including those of the bloodstream which can become fatal. Originally treated by penicillin, Staphylococcus aureus infections quickly developed resistance to this antibiotic as well as almost all other beta-lactam antibiotics. When Staphylococcus aureus developed resistance to the antibiotic methicillin through the mecA gene (located on a mobile genetic element) the bacterium became a major concern for its spread especially in hospitals as well as the difficulty of treating the now methicillin-resistant infections (MRSA infections). An improved effort on maintaining proper hand hygiene in hospitals has been an effective way of decreasing the spread of MRSA in hospitals (hand-to-hand contact is a common method of transferring the bacteria to a new host). With the introduction of the procedure of screening newly admitted patients in hospitals, the spread of MRSA in hospitals can be further reduced as it allows for the quick isolation of MRSA colonized patients (preventing contact and spread). The use of new antibiotic treatment plans however has not been as a successful as one would hope in killing/getting rid of MRSA infections. As drugs or antibiotics are used repeatedly to treat the bacterial infection, it is only a matter of time before resistance to these treatments is also developed. This is a fear associated with the treatment of MRSA infections with vancomycin as some resistance strains are emerging through the presence of a vanA gene. It is also possible that resistance to certain antibiotics by Staphylococcus aureus can be spread to other bacteria infecting a host at the same time. The uptake of new genes through plasmids is a concern for the spread of these resistances. For these reasons, the research currently being done with naturally occurring substances that have the ability to “revive” the effectiveness of antibiotics is very exciting and promising. It is even likely that these synergistic treatments can be used on other bacterial infections that are resistant to a variety of antibiotics.