Infection Mechanism of Genus Ebolavirus: Difference between revisions
| Line 50: | Line 50: | ||
So far, most of the processes by which Ebola escapes or hampers immune response involve structural proteins on the virus itself. However, one of the most important, and most misunderstood, proteins is the sGP protein that is released by Ebola into the bloodstream. During viral infection, the stability of the vascular endothelium is destroyed by proinflammatory cytokines released from hijacked macrophages or by the cytoxic effects of Ebola replication. However, when sGP is present, there is an apparent recovery of endothelial cell barrier function. While this seems contrary to the actions of other viral proteins, it actually prevents neutrophils and other white blood cells from moving from the blood stream to areas of infection, preventing a coordinated immune response (Wahl-Jensen et al. 2005). | So far, most of the processes by which Ebola escapes or hampers immune response involve structural proteins on the virus itself. However, one of the most important, and most misunderstood, proteins is the sGP protein that is released by Ebola into the bloodstream. During viral infection, the stability of the vascular endothelium is destroyed by proinflammatory cytokines released from hijacked macrophages or by the cytoxic effects of Ebola replication. However, when sGP is present, there is an apparent recovery of endothelial cell barrier function. While this seems contrary to the actions of other viral proteins, it actually prevents neutrophils and other white blood cells from moving from the blood stream to areas of infection, preventing a coordinated immune response (Wahl-Jensen et al. 2005). | ||
== | ==Viral Entry== | ||
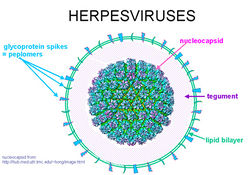
[[Image:Herpes Virus.jpg|thumb|250px|right|The structure of a general alphaherpesvirus. [http://www.everclr3.net/facts/hsv.html].]] | [[Image:Herpes Virus.jpg|thumb|250px|right|The structure of a general alphaherpesvirus. [http://www.everclr3.net/facts/hsv.html].]] | ||
Endocytosis offers an efficient way for viruses to cross the physical barrier of the plasma membrane and move throughout the underlying matrix to cytoplasmic sites suitable for replication. The entry pathway of Ebolavirus must be elucidated if viral pathogenicity is to be understood and drugs are to be developed. After binding to the host cell receptor, the internalized virus is transported through successive endocytic vesicles until it reaches a compartment where conditions favor a conformation shift in viral GP that causes membrane fusion. While there is much evidence suggesting that Ebolavirus enters cells through pH-dependent endocytosis (Schornberg et al. 2006), the specific mechanism has not yet been clearly defined. | |||
Previous studies to clarify the entry have produced conflicting results, with involvement of clathrin-mediated endocytosis (Bhattacharyya et al. 2010), caveolin-mediated endocytosis (Empig & Goldsmith 2002) and a Rho GTPase-dependent pathway that may suggest macropinocytosis all being implicated (Quinn et al. 2009). However, all of these experiments were performed using retrovirus-based pseudotypes in which GP is coated onto the surface of a retrovirus capsid containing a recombinant genome. This removes the need to work in a biosafety level four laboratory, but suffers from non-ideal biochemical characteristics. Only recently have experiments been performed with wild-type Ebolavirus Zaire that demonstrate that cellular entry involves uptake by a macropinocytosis-like mechanism (Saeed et al. 2010). | |||
<b><font color = red>TALK ABOUT MACROPINOCYTOSIS AND ENTRY. | |||
CONNECTION FROM ENTRY TO TRANSCRIPTION AND REST OF CYCLE.</b></font> | |||
==Reproductive Cycle== | ==Reproductive Cycle== | ||
Revision as of 02:22, 3 November 2010
A Viral Biorealm page on the family Infection Mechanism of Genus Ebolavirus
Ebola hemorrhagic fever is a severe, often fatal, disease in humans and non-human primates caused by Ebolavirus. Even though the virus first appeared in the Democratic Republic of the Congo (formerly Zaire) in the summer of 1976 and has only sporadically reemerged, it has still managed to capture the attention of the world due to death rates that can be as high as 90% as well as the visceral manner in which it kills (CDC 2010, Johnson & Breman 1978). Few viruses have the ability to turn internal organs into a soup that promptly flows out of the body, so those that do tend to capture the public eye.
Despite the low incidence of infections, the lethality and potential airborne transmission of Ebolavirus makes it a severe biological threat. This is compounded by the possibility of the virus being obtained from the wild for use as a biological weapon (Borio et al. 2002). As of present, there are no licensed vaccines or specific antiviral treatments available for Ebolavirus infections, with significant progress only made in the development of vaccines for nonhuman primates (Pratt et al. 2010). Therefore, the elucidation of the viral replication cycle as well as complete understanding of viral proteins is necessary for the production of human vaccines.
Ebola Hemorrhagic Fever
One of the reasons that Ebola is so dangerous is that its symptoms are varied and appear quickly, yet are close enough to those of other viruses that Ebola is not rapidly recognized. Initial symptoms include high fever, severe headache, severe weakness, exhaustion, sore throat, nausea, dizziness and muscle, joint or abdominal pain. The unfortunate thing about these symptoms is that they are easily mistaken for malaria, typhoid fever, dysentery, influenza or various bacterial infections, all of which are far more common in regions where Ebola is prevent, but also less fatal (CDC 2010).
After this period, the fever often progresses to cause more serious symptoms: diarrhea, dark or bloody feces, vomiting blood, red eyes due to distention and hemorrhage of sclerotic arterioles, petechia, maculopapular rash and purpura. There is often gastrointestinal bleeding from the mouth and rectum, sometimes leading to the sloughing of the gut and venting from the anus. Internal and external hemorrhage from orifices, such as the nose and mouth, may also occur. As the virus progresses, bleeding in the brain can lead to severe depression, seizures and delirium (CDC 2010).
The span of time from onset of symptoms to death is usually between 7 and 14 days. By the second week of the infection, the patient will either experience a full recovery or undergo systemic multi-organ failure. Mortality rates are generally high, ranging from 50-90% depending on the specific strain. The cause of death is normally due to hypovolemic shock or organ failure (WHO 2010).
Virion Structure
Filoviridae, of which Ebolavirus is a member, is a family of viruses that contain single, linear, negative-sense ssRNA genomes. The family name was derived from the Latin word filum, which alludes to the thread-like appearance of the virions when viewed under an electron microscope. Filoviruses have been divided into two genera: Ebola-like viruses with species Zaire, Sudan, Reston, Cote d’Ivoire and Bundibugyo; and Marburg-like viruses with the single species Marburg. All of these are responsible for hemorrhagic fevers in primates that are characterized by often fatal bleeding and coagulation abnormalities (Klenk & Feldmann 2004).
The tubular Ebola virions are generally 80 nm in diameter and 800 nm long. In the center of the particle is the viral nucleocapsid which consists of the helical ssRNA genome wrapped about the NP, VP35, VP30 and L proteins. This structure is then surrounded by an outer viral envelope derived from the host cell membrane that is studded with 10 nm long viral glycoprotein (GP) spikes. Between the capsid and envelope are viral proteins VP40 and VP24 (Feldmann et al. 1993).
The genome of each virion is around 19kb in length, and codes for seven structural and one non-structural protein. The gene order is as follows: 3′ – leader – NP – VP35 – VP40 – GP/sGP – VP30 – VP24 – L – trailer – 5′. The leader and trailer regions are not transcribed, but carry important signals that control transcription, replication and packaging of the genome into new virions (Crary et al. 2003). The seven genes each consist of their respective open reading frame as well as long non-translated sequences of unknown purpose that flank the coding regions (Klenk & Feldmann 2004).
The envelope GP spike is the only viral surface protein, and thus has been implicated in cellular entry by mediating receptor binding and membrane fusion. It has also been shown as the crucial factor for Ebolavirus pathogenicity (Yonezawa et al. 2005). The product of the third gene, VP40, is located beneath the viral envelope where it helps to maintain structural integrity. It has also been associated with late endosomes and likely mediates filovirus budding due to its ability to induce its own release from cells in the absence of all other viral proteins (Jasenosky et al. 2001).
The second matrix protein, VP24, has been shown to suppress interferon production, and may also be important in regulation prior to budding. However, interferon interference is not the only function of VP24. Other experiments have shown that this protein, along with VP35 and NP are sufficient to form nucleocapsid structures (Huang et al. 2002). Lastly, VP24 is necessary for the correct assembly of a functional nucleocapsid, as lack of VP24 leads to reduced transcription and/or translation of VP30 (Hoenen et al. 2006).
The remaining structural proteins form the nucleocapsid, and are thus intimately associated with the viral genome. These are the nucleoprotein NP, the polymerase cofactor VP35, the viral-specific transcription activator VP30 and the viral RNA polymerase L. These nucleocapsid proteins have a dual function in the viral replication cycle: they are involved as structural components, but also catalyze replication and transcription of the genome. While NP, VP35 and L are sufficient for replication, transcription initiation will not proceed without VP30 (Mühlberger et al. 1999).
Ebolavirus genomes code for an additional protein, the non-structural secreted GP (sGP). This truncated protein is also encoded by the fourth gene, but synthesis is due to translation from non-edited mRNA species. The structural form requires a reading frame shift or transcriptional editing of a single nucleotide. Therefore, almost 80% of the proteins encoded by this gene are in the sGP form (Simmons et al. 2002). This protein is not found in virus particles, but is instead secreted from infected cells into the blood (Volchkov et al. 1995).
NEED TO TALK MORE ABOUT GP AND sGP
Immune System Evasion
One of the reasons that Ebola is so deadly is that it has multiple ways of interfering with or avoiding the human immune system. While the virus is busy destroying the human body, the immune system is either still in the process of discovering that there is a problem, or is in such disarray that it would be next to impossible to mobilize a unified effort to fight off the invader. This virus is able to hijack antibodies, take control of macrophages, inhibit movement of neutrophils, block interferon and destroy dendritic cells.
Antibodies normally function by attaching to their respective antigen, then starting a complement cascade that ultimately leads to the pathogen’s destruction by phagocytosis. After two IgG antibodies bind the GP spikes, complement component one (C1) binds the complex via the Fc portion of the antibodies. This complex then attaches to a C1q ligand on the surface of cells. While this normally promotes endocytosis of the target antigen by intracellular signaling, in this case it only serves in the binding of Ebola to viral receptors. C1q ligands have been identified on many cell types, including macrophages, the primary target of the Ebola virus. Therefore, while antibodies normally protect the body, this virus is able to use them for faster and easier attachment to target cells (Takada et al. 2003).
In addition being an essential cofactor for the viral RNA polymerase complex, VP35 has been identified as an inhibitor of multiple components of the interferon (IFN) pathways. These are a family of cytokines produced in response to viral infection and regulated by many transcription factors, including interferon regulatory factor 3 (IRF-3). After IRF-3 is stimulated by dsRNA or viral infection, IFN-α/β are secreted. They activate the Janus tyrosine kinase / signal transducer and activator (JAK/STAT) signaling pathway that stimulates transcription of genes encoding antiviral proteins such as protein kinase R (PKR). Upon binding to dsRNA, PKR is activated and phosphorylates translation initiation factor eIF-2, which then arrests protein synthesis and inhibits viral replication (Feng et al. 2007).
VP35 also blocks PKR activation via its C-terminal IRF inhibitory domain, and can even go so far as to reverse PKR phosphorylation (Schümann 2009). FOCUS MORE ON VP35 AND THIS PATH.
As previously mentioned, the second matrix protein, VP24, suppresses interferon production. The binding of IFN-α/β and IFN-γ to their respective receptors activates STAT complexes, a family of transcription factors that regulate immune system gene expression. Normally, STAT1:STAT2 heterodimers and STAT1 homodimers are transported to the nucleus by karyopherin α1 and activate numerous genes involved in antiviral activities. However, VP24 competes with STAT1 to bind karyopherin α1, blocking nuclear accumulation and leading to inhibition of IFN signaling (Reid 2006).
During infection, Ebola controlled macrophages release nitric oxide (NO), a gaseous hormone that normally functions in cell communication. However, in high concentrations, NO depresses the mitochondrial membrane potential, which causes apoptosis in NK and other nearby cells. This is yet another way that Ebola disrupts the immune system (Geisbet et al. 2003).
So far, most of the processes by which Ebola escapes or hampers immune response involve structural proteins on the virus itself. However, one of the most important, and most misunderstood, proteins is the sGP protein that is released by Ebola into the bloodstream. During viral infection, the stability of the vascular endothelium is destroyed by proinflammatory cytokines released from hijacked macrophages or by the cytoxic effects of Ebola replication. However, when sGP is present, there is an apparent recovery of endothelial cell barrier function. While this seems contrary to the actions of other viral proteins, it actually prevents neutrophils and other white blood cells from moving from the blood stream to areas of infection, preventing a coordinated immune response (Wahl-Jensen et al. 2005).
Viral Entry
Endocytosis offers an efficient way for viruses to cross the physical barrier of the plasma membrane and move throughout the underlying matrix to cytoplasmic sites suitable for replication. The entry pathway of Ebolavirus must be elucidated if viral pathogenicity is to be understood and drugs are to be developed. After binding to the host cell receptor, the internalized virus is transported through successive endocytic vesicles until it reaches a compartment where conditions favor a conformation shift in viral GP that causes membrane fusion. While there is much evidence suggesting that Ebolavirus enters cells through pH-dependent endocytosis (Schornberg et al. 2006), the specific mechanism has not yet been clearly defined.
Previous studies to clarify the entry have produced conflicting results, with involvement of clathrin-mediated endocytosis (Bhattacharyya et al. 2010), caveolin-mediated endocytosis (Empig & Goldsmith 2002) and a Rho GTPase-dependent pathway that may suggest macropinocytosis all being implicated (Quinn et al. 2009). However, all of these experiments were performed using retrovirus-based pseudotypes in which GP is coated onto the surface of a retrovirus capsid containing a recombinant genome. This removes the need to work in a biosafety level four laboratory, but suffers from non-ideal biochemical characteristics. Only recently have experiments been performed with wild-type Ebolavirus Zaire that demonstrate that cellular entry involves uptake by a macropinocytosis-like mechanism (Saeed et al. 2010).
TALK ABOUT MACROPINOCYTOSIS AND ENTRY.
CONNECTION FROM ENTRY TO TRANSCRIPTION AND REST OF CYCLE.
Reproductive Cycle
Again, very little is known about the specific reproductive cycle of FPTHV, though generalizations can be made after studying similar alphaherpesviruses. These virions make initial contact with host cells through the interactions of envelope glycoproteins gB and/or gC with the surface glycosaminoglycan heparan sulfate. While this binding is not essential for infection, it can increase entry efficiency by concentrating the virus on the cell surface so viral ligand gD can find cell receptors [21].
Viral glycoproteins gB, gD, gH and gL then act together to induce fusion of the envelope with the cell membrane following association with one of three classes of entry receptors: herpesvirus entry mediator, a member of the tumor necrosis factor receptor family; nectin-1 and nectin-2, members of the immunoglobulin family; and specific sites in heparin sulfate [22]. The virion moves to the nucleus and attaches to a nuclear entry pore, then inserts its DNA through the capsid portal. Each icosahedral virion contains one ring-shaped capsid portal, which is formed by twelve copies of the portal protein UL6 [4].
In the nucleus, the DNA circularizes and uses host enzymes to code for its “early genes.” The mRNA migrates to the cytoplasm and codes for proteins that then must return to the nucleus in order to help with further transcription. Late genes then code for capsid subunits, as well as for proteins that are incorporated into the nuclear cell membrane to become part of the primary viral envelope. In the meantime, the viral DNA continues to replicate in a rolling-circle mechanism that rapidly produces end-to-end copies called concatemers that must be cut into individual genomes as they are packaged into capsids [15].
The first step in nuclear egress of alphaherpesviruses is a budding process at the inner leaflet of the nuclear membrane in which capsids acquire a primary envelope [17]. The UL31 and UL34 proteins are necessary for efficient exit, with the UL34 protein inducing protein kinase C to soften the nuclear lamina via phosphorylation [20]. Following this, the primary envelope fuses with the outer leaflet of the nuclear membrane and releases the viral capsid, sans primary envelope and primary tegument, into the cytoplasm [6].
Once there, the final tegument is formed. These proteins help link capsid subunits to the cytoplasmic tails of envelope glycoproteins to secure the integrity of the virion. The first layer of tegument around the capsid is icosahedrally shaped and composed of UL36. A second layer is composed of UL37, while UL48 appears critical for tegument assembly [18]. After this protein coat is assembled, tegumented capsides bud into trans-Golgi vesicles to form a final envelope. Tegument protein UL49 interacts with the cytoplasmic tails of vesicle membrane glycoproteins gE/I and gM to help drive this last budding process [17].
Like many other herpes viruses, FPTHV can also persist as a latent infection. It expresses Latency Associated Transcript RNA that regulates the host genome and interferes with the natural cell death mechanism, ensuring that a reservoir of virus is ready for another lytic outbreak [17].
Viral Ecology & Pathology
Skin tumors in turtles affected by FP are characterized by a branching collagen matrix mixed with scattered fibroblasts covered by an epidermis that is abnormally thickened [11]. Cell dependent levels of FPTHV pol DNA suggest that lytic viral production occurs in keratinocytes, while the majority of FPTHV viral genomes are contained in fibroblasts either as an episome, a portion of genetic material that can exist independently of the main genome, or as part of host DNA [7]. This genome sequence has also been used to prove that FPTHV plays a dominant role in the maintenance of the tumor state, despite the fact that infection appears predominantly latent in fibropapilloma cells [8,19].
It has been suggested that FPTHV infection occurs when marine turtles return to their natal breeding grounds [27]. Recently, marine leeches of the genus Ozobranchus have been associated with FPTHV DNA, and thus considered candidate vectors. However, no viral pol transcripts were found in the leech RNA samples, showing that the leech vector role is most likely limited to mechanical transmission between turtles [8]. This does not rule out waterborne infection though, as the lung-eye-trachea virus, another herpesvirus of marine turtles, is incredibly stable in salt water [5].
References
Basler, C.F., Wang, X., Mühlberger, E., Volchkov, V., Paragas, J., Klenk, H.D., Garcia-Sastre, A. & Palese, P. (2000). The Ebola virus VP35 protein functions as a type I IFN antagonist. PNAS, 97, 12289-12294.
Basler, C.F., Mikulasova, A., Martinez-Sobrido, L., Paragas, J., Mühlberger, E., Bray, M., Klenk, H.D., Palese, P. & García-Sastre, A. (2003). The Ebola Virus VP35 Protein Inhibits Activation of Interferon Regulatory Factor 3. Journal of Virology, 77(14), 7945-7956.
Bavari, S., Bosio, C.M., Wiegand, E., Ruthel, G., Will, A.B., Geisbert, T.W., Hevey, M., Schmaljohn, C., Schmaljohn, A. & Aman, M.J. (2002). Lipid Raft Microdomains: A Gateway for Compartmentalized Trafficking of Ebola and Marburg Viruses. The Journal of Experimental Medicine, 195(5), 593-602.
Bhattacharyya, S., Warfield, K.L., Ruthel, G., Bavari, S., Aman, M.J. & Hope, T.J. (2010). Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology, 401, 18-28.
Borio, L., Inglesby, T., Peters, C.J., Schmaljohn, A.L., Hughes, J.M., Jahrling, P.B., Ksiazek, T., Johnson, K.M., Meyerhoff, A., O’Toole, T., Ascher, M.S., Bartlett, J., Breman, J.G., Eitzen, Jr., E.M., Hamburg, M., Hauer, J., Henderson, D.A., Johnson, R.T., Kwik, G., Layton, M., Lillibridge, S., Nabel, G.J., Osterholm, M.T., Perl, T.M., Russell, P. & Tonat, K. (2002). Hemorrhagic fever viruses as biological weapons: medical and public health management. Journal of the American Medical Association, 287, 2391-2405.
CDC Special Pathogens Branch. (2010). Ebola Hemorrhagic Fever Case Count and Location List. Ebola Hemorrhagic Fever Information Packet.
Cheng, P.C., Cherukuri, A., Dykstra, M., Malapati, S., Sproul, T., Chen, M.R. & Pierce, S.K. (2001). Floating the raft hypothesis: the roles of lipid rafts in B cell antigen receptor function. Semininars in Immunology, 13, 107-114.
Crary, S.M., Towner, J.S., Honig, J.E., Shoemaker, T.R. & Nichol, S.T. (2003). Analysis of the role of predicted RNA secondary structures in Ebola virus replication. Virology, 306(2), 210-218.
Emerson, S.U. (1982). Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell, 31, 635-642.
Empig, C.J. & Goldsmith, M.A. (2002). Association of the caveola vesicular system with cellular entry by filoviruses. Journal of Virology, 76, 5266-5270.
Feldmann, H., Klenk, H.D. & Sanchez, A. (1993). Molecular biology and evolution of filoviruses. Archives of Virology, Supplementum, 7, 81-100.
Hoenen, T., Groseth, A., Kolesnikova, L., Theriault, S., Ebihara, H., Hartlieb, B., Bamberg, S., Feldmann, H., Ströher, U. & Becker, S. (2006). Infection of Naïve Target Cells with Virus-Like Particles: Implications for the Function of Ebola Virus VP24. Journal of Virology, 80(14), 7260-7264.
Huang, Y., Xu, L., Sun, Y. & Nabel, G.J. (2002). The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Molecular Cell, 10, 307-316.
Iverson, L.E. & Rose, J.K. (1982). Sequential synthesis of 5’-proximal vesicular stomatitis virus mRNA sequences. Journal of Virology, 44, 356-365.
Jasenosky, L.D., Neumann, G., Lukashevich, I. & Kawaoka, Y. (2001). Ebola Virus VP40-Induced Particle Formatin and Association with the Lipid BIlayer. Journal of Virology, 75(11), 5205-5214.
John, S.P., Wang, T., Steffen, S., Longhi, S., Schmaljohn, C.S. & Jonsson, C.B. (2007). Ebola Virus VP30 Is an RNA Binding Protein. Journal of Virology, 81(17), 8967-8976.
Johnson, K.M. & Breman, J.G. (1978) Ebola haemorrhagic fever in Zaire, 1976. Bulletin of the World Health Organization, 56(2), 271-293.
Klenk, H.-D. & Feldmann, H. (2004). Ebola and Marburg Viruses: Molecular and Cellular Biology. Wymondham: Horizon Bioscience.
Martínez, M.J., Biedenkopf, N., Volchkova, V., Hartlieb, B., Alazard-Dany, N., Reynard, O., Becker, S. & Volchkov, V. (2008). Role of Ebola Virus VP30 in Transcription Reinitiation. Journal of Virology, 82(24), 12569-12573.
Modrof, J., Becker, S. & Muhlberger, E. (2003). Ebola virus transcription activator VP30 is a zinc-binding protein. Journal of Virology, 77, 3334-3338.
Moran, M. & Miceli, M.C. (1998). Engagement of GPI-linked CD48 contributes to TCR signals and cytoskeletal reorganization: a role for lipid rafts in T cell activation. Immunity, 9, 787-796.
Mühlberger, E.,Weik, M., Volchkov, V.E., Klenk, H.D. & Becker, S. (1999). Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. Journal of Virology, 73, 2333-2342.
Pratt, W.D., Wang, D., Nichols, D.K., Luo, M., Woraratanadharm, J., Dye, J.M., Holman, D.H. & Dong, J.Y. (2010). Protection of Nonhuman Primates against Two Species of Ebola Virus Infection with a Single Complex Adenovirus Vector. Clinical and Vaccine Immunology, 17(4), 572-581.
Quinn, K., Brindley, M.A., Weller, M.L., Kaludov, N., Kondratowicz, A., Hunt, C.L., Sinn, P.L., McCray, P.B., Stein, C.S., Davidson, B.L., Flick, R., Mandell, R., Staplin, W., Maury, W. & Chiorini, J.A. (2009). Rho GTPases Modulate Entry of Ebola Virus and Vesicular Stomatitis Virus Pseudotyped Vectors. Journal of Virology, 83(19), 10176-10186.
Reid, S.P., Leung, L.W., Hartman, A.L., Martinez, O., Shaw, M.L., Carbonnelle, C., Volchkov, V.E., Nichol, S.T. & Basler, C.F. (2006). Ebola Virus VP24 Binds Karyopherin α1 and Blocks STAT1 Nuclear Accumulation. Journal of Virology, 80(11), 5156-5167.
Saeed, M.F., Kolokoltsov, A.A., Albrecht, T. & Davey, R.A. (2010). Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes. PLOS Pathogens, 6(9).
Saifuddin, M., Hedayati, T., Atkinson, J.P., Holguin, M.H., Parker, C.J. & Spear, G.T. (1997). Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. Journal of General Virology, 78, 1907-1911.
Sanchez, A., Kiley, M.P., Holloway, B.P. & Auperin, D.D. (1993). Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Research, 29, 215-240.
Schümann, M., Gantke, T. & Mühlberger, E. (2009). Ebola Virus VP35 Antagonizes PKR Activity through Its C-Terminal Interferon Inhibitory Domain. Journal of Virology, 83(17), 8993-8997.
Schornberg, K., Matsuyama, S., Kabsch, K., Delos, S., Bouton, A. & White, J. (2006). Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. Journal of Virology, 80(8), 4174-4178.
Volchkov, V.E., Becker, S., Vochkova, V.A., Ternovoj, V.A., Kotov, A.N., Netesov, S.V. & Klenk, H.D. (1995). GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology, 214, 421-430.
Watanabe, S., Noda, T., Halfmann, P., Jasenosky, L. & Kawaoka, Y. (2007). Ebola Virus (EBOV) VP24 Inhibits Transcription and Replication of the EBOV Genome. The Journal of Infectious Diseases, 196, S284-290.
Weik, M., Modrof, J., Klenk, H.D., Becker, S. & Mühlberger, E. (2002). Ebola Virus VP30-Mediated Transcription Is Regulated by RNA Secondary Structure Formation. Journal of Virology, 76(17), 8532-8539.
World Health Organization. (2010). Ebola Hemorrhagic Fever. Epidemic and Pandemic Alert and Response (EPR).
Yonezawa, A., Cavrois, M. & Greene, W.C. (2005). Studies of Ebola Virus Glycoprotein-Mediated Entry and Fusion by Using Pseudotyped Human Immunodeficiency Virus Type 1 Virions: Involvement of Cytoskeletal Proteins and Enhancement by Tumor Necrosis Factor Alpha. Journal of Virology, 79(2), 918-926.
Page authored for BIOL 375 Virology, September 2010
