Infection Mechanism of Genus Ebolavirus
A Viral Biorealm page on the family Infection Mechanism of Genus Ebolavirus
Ebola hemorrhagic fever is a severe, often fatal, disease in humans and non-human primates caused by Ebolavirus. Even though the virus first appeared in the Democratic Republic of the Congo (formerly Zaire) in the summer of 1976 and has only sporadically reemerged, it has still managed to capture the attention of the world due to death rates that can be as high as 90% as well as the visceral manner in which it kills (CDC 2010, Johnson & Breman 1978). Few viruses have the ability to turn internal organs into a soup that promptly flows out of the body, so those that do tend to capture the public eye.
Despite the low incidence of infections, the lethality and potential airborne transmission of Ebolavirus makes it a severe biological threat. This is compounded by the possibility of the virus being obtained from the wild for use as a biological weapon (Borio et al. 2002). As of present, there are no licensed vaccines or specific antiviral treatments available for Ebolavirus infections, with significant progress only made in the development of vaccines for nonhuman primates (Pratt et al. 2010). Therefore, the elucidation of the viral replication cycle as well as complete understanding of viral proteins is necessary for the production of human vaccines.

Virus Classification
Group: Group 1 (dsDNA viruses)
Order: Herpesvirales
Family: Herpesviridae
Subfamily: Alphaherpesvirinae
Genus: As-Yet-Uncharacterized
Since fibropapilloma-associated turtle herpesvirus (FPTHV) provides a new model system for studying virus-related oncogenesis, its genome was partially sequenced in 2004 as basis for further study. While regions of the FPTHV polymerase (pol) gene showed 98% sequence homology with nine highly conserved herpesvirus pol domains, there was significant variation in alphaherpesvirus-specific consensus (7% not similar). This confirmed that the virus is a member of Alphaherpesvirinae, but it does not appear to belong to the four existing genera. This study also revealed the presence of at least four FPTHV variants that are separated by geography, and not by the species of the host [9].
Description and Significance
Infection by FPTHV has been associated with fibropapillomatosis (FP), a neoplastic disease of marine turtles [14]. This condition manifests itself through the presence of epithelial fibropapillomas, tumors consisting of the epidermal and dermal skin layers, as well as internal fibromas, benign tumors composed of fibrous or connective tissue [10]. Although the tumors are considered benign, the disease can be life-threatening; large external fibropapillomas interfere with vision and feeding, while visceral fibromas can interfere with organ function [13].
Even though first reports of infection were only in green turtles (Chelonia mydas), cases have recently been confirmed in both loggerhead (Caretta caretta) and olive ridley (Lepidochely olivacea) species [26]. In the 1990s, it was believed that FP would be responsible for the extinction of marine turtles. However, recent studies have shown that wild turtles can, and will, recover from the disease [3]. Despite this fact, the incidence of FP has gradually increased in recent times, providing an additional ecological stress on magnificent creatures whose long-term survival is already in doubt.
Genome Structure
The genome of FPTHV consists of linear, dsDNA with unpaired, complementary nucleotides at each terminus. While the entire genome has not yet been sequenced, it is expected to be between 124 and 241 kbp, based on the size of fully sequenced alphaherpesviruses [9]. Overall, the sequence resembles that of another alphaherpesvirus, herpes simplex virus, which can be responsible for genital herpes [1]. Thus, FPTHV most likely contains at least 74 genes arranged in two unique regions, the long unique region (UL) and the short unique region (US), flanked by inverted repeats [16,24].
Protein coding genes are transcribed by host RNA polymerase II [2]. The first genes to be expressed during infection, immediate early genes, encode enzymes involved in nucleotide metabolism, DNA replication and for glycoproteins of the viral envelope. In contrast, late genes code for proteins of the viral capsid [12].
Virion Structure
The exact structure of the FPTHV virion has not been specifically determined. However, much insight can be gained by examining other alphaherpesviruses since the genomes show great homology [9].
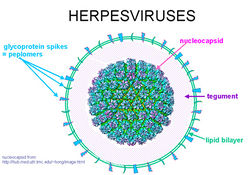
Alphaherpesvirus virions are composed of a large, dsDNA genome enclosed in an icosahedral capsid. This is coated first in a layer of tegument proteins and then in an envelope composed of roughly a dozen viral proteins and glycoproteins. The capsid is 125 nm in diameter and consists of four main structural proteins, VP5, VP19C, VP23 and VP26, along with a number of minor capsid proteins. A number of these minor subunits are required for the packing of replicated viral DNA into new capsid shells [25]. The viral envelope is derived from vesicles of the trans-Golgi network, and contains several virus-encoded glycoproteins that are absolutely essential for viral entry, including gB, gH and gL [23].
Reproductive Cycle
Again, very little is known about the specific reproductive cycle of FPTHV, though generalizations can be made after studying similar alphaherpesviruses. These virions make initial contact with host cells through the interactions of envelope glycoproteins gB and/or gC with the surface glycosaminoglycan heparan sulfate. While this binding is not essential for infection, it can increase entry efficiency by concentrating the virus on the cell surface so viral ligand gD can find cell receptors [21].
Viral glycoproteins gB, gD, gH and gL then act together to induce fusion of the envelope with the cell membrane following association with one of three classes of entry receptors: herpesvirus entry mediator, a member of the tumor necrosis factor receptor family; nectin-1 and nectin-2, members of the immunoglobulin family; and specific sites in heparin sulfate [22]. The virion moves to the nucleus and attaches to a nuclear entry pore, then inserts its DNA through the capsid portal. Each icosahedral virion contains one ring-shaped capsid portal, which is formed by twelve copies of the portal protein UL6 [4].
In the nucleus, the DNA circularizes and uses host enzymes to code for its “early genes.” The mRNA migrates to the cytoplasm and codes for proteins that then must return to the nucleus in order to help with further transcription. Late genes then code for capsid subunits, as well as for proteins that are incorporated into the nuclear cell membrane to become part of the primary viral envelope. In the meantime, the viral DNA continues to replicate in a rolling-circle mechanism that rapidly produces end-to-end copies called concatemers that must be cut into individual genomes as they are packaged into capsids [15].
The first step in nuclear egress of alphaherpesviruses is a budding process at the inner leaflet of the nuclear membrane in which capsids acquire a primary envelope [17]. The UL31 and UL34 proteins are necessary for efficient exit, with the UL34 protein inducing protein kinase C to soften the nuclear lamina via phosphorylation [20]. Following this, the primary envelope fuses with the outer leaflet of the nuclear membrane and releases the viral capsid, sans primary envelope and primary tegument, into the cytoplasm [6].
Once there, the final tegument is formed. These proteins help link capsid subunits to the cytoplasmic tails of envelope glycoproteins to secure the integrity of the virion. The first layer of tegument around the capsid is icosahedrally shaped and composed of UL36. A second layer is composed of UL37, while UL48 appears critical for tegument assembly [18]. After this protein coat is assembled, tegumented capsides bud into trans-Golgi vesicles to form a final envelope. Tegument protein UL49 interacts with the cytoplasmic tails of vesicle membrane glycoproteins gE/I and gM to help drive this last budding process [17].
Like many other herpes viruses, FPTHV can also persist as a latent infection. It expresses Latency Associated Transcript RNA that regulates the host genome and interferes with the natural cell death mechanism, ensuring that a reservoir of virus is ready for another lytic outbreak [17].
Viral Ecology & Pathology
Skin tumors in turtles affected by FP are characterized by a branching collagen matrix mixed with scattered fibroblasts covered by an epidermis that is abnormally thickened [11]. Cell dependent levels of FPTHV pol DNA suggest that lytic viral production occurs in keratinocytes, while the majority of FPTHV viral genomes are contained in fibroblasts either as an episome, a portion of genetic material that can exist independently of the main genome, or as part of host DNA [7]. This genome sequence has also been used to prove that FPTHV plays a dominant role in the maintenance of the tumor state, despite the fact that infection appears predominantly latent in fibropapilloma cells [8,19].
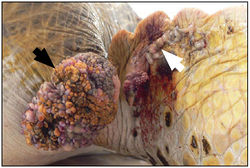
It has been suggested that FPTHV infection occurs when marine turtles return to their natal breeding grounds [27]. Recently, marine leeches of the genus Ozobranchus have been associated with FPTHV DNA, and thus considered candidate vectors. However, no viral pol transcripts were found in the leech RNA samples, showing that the leech vector role is most likely limited to mechanical transmission between turtles [8]. This does not rule out waterborne infection though, as the lung-eye-trachea virus, another herpesvirus of marine turtles, is incredibly stable in salt water [5].
References
[1] Ackermann, M. (2010). Characterization of the Fibropapilloma-associated Marine Turtle Herpesvirus. Retrieved 9/6/10 from <http://www.forschungsportal.ch/unizh/p6785.htm>.
[2] Alwine, J.C., Steinhart, W.L. & Hill, C.W. (1974). Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology, 60, 302–307.
[3] Bennett, P., Keuper-Bennett, U. & Balazs, G.H. (1999). Photogrphic evidence for the regression of fibropapillomas afflicting green turtles at Honokowai, Maui, in the Hawaiian Islands. 19th Annual Sea Turtle Symposium.
[4] Cardone, G., Winkler, D.C., Trus, B.L., Cheng, N., Heuser, J.E., Newcomb, W.W., Brown, J.C. & Steven, A.C. (2007). Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology, 361(2), 426-434.
[5] Curry, S.S., Brown, D.R., Gaskin, J.M., Jacobson, E.R., Ehrhart, L.M., Blahak, S., Herbst, L.H. & Klein, P.A. (2000). Persistent infectivity of a disease-associated herpesvirus in green turtles after exposure to seawater. J. Wildl. Dis., 36, 792-797.
[6] Enquist, L.W., Husak, P.J., Banfield, B.W. & Smith, G.A. (1999). Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res., 51, 237–347.
[7] Ganem, D. (2006). KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol., 1, 273-296.
[8] Greenblatt, R.J., Work, T.M., Balazs, G.H., Sutton, C.A., Casey, R.N. & Casey, J.W. (2004). The Ozobranchus leech is a candidate mechanical vector for the fibropapilloma-associated turtle herpesvirus found latently infecting skin tumors on Hawaiian green turtles (Chelonia mydas). J. Virol., 321, 101-110.
[9] Greenblatt, R.J., Quackenbush, S.L., Casey, R.N., Rovnak, J., Balazs, G.H., Work, T.M., Casey, J.W. & Sutton, C.A. (2005). Genomic Variation of the Fibropapilloma-Associated Marine Turtle Herpesvirus across Seven Geographic Areas and Three Host Species. J. Virol., 79(2), 1125-1132.
[10] Herbst, L.H. (1994). Fibropapillomatosis of marine turtles. Annu. Rev. Fish Dis., 4, 389-425.
[11] Herbst, L.H., Jacobson, E.R., Klein, P.A., Balazs, G.H., Moretti, R., Brown, T. & Sundberg, J.P. (1999). Comparative pathology and pathogenesis of spontaneous and experimentally induced fibropapillomas of green turtles (Chelonia mydas). Vet. Pathol., 36, 551–564.
[12] Honess, R.W. & Roizman, B. (1974). Regulation of herpesvirus macromolecular synthesis. I. cascade regulation of the synthesis of three groups of viral proteins. J. Virol., 14, 8–19.
[13] Jacobson, E.R., Mansell, J.L., Sundberg, J.P., Hajjar, L., Reichmann, M.E., Ehrhart, L.M.,Walsh, M. & Murru F. (1989). Cutaneous fibropapillomas of green turtles (Chelonia mydas). J. Comp. Pathol., 101, 39–52.
[14] Lackovich, J.K., Brown, D.R., Homer, B.L., Garber, R.L., Mader, D.R., Moretti, R.H., Patterson, A.D., Herbst, L.H., Oros, J., Jacobson, E.R., Curry, S.S. & Klein, P.A. (1999). Association of herpesvirus with fibropapillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta caretta in Florida. Dis. Aquat. Org., 37, 889–897.
[15] Lehman, I.R. & Boehmer, P.E. (1999). Replication of Herpes Simplex Virus DANN. J. Biol. Chem., 274, 28059-28062.
[16] McGeoch, D.J., Rixon, F.J. & Davison, A.J. (2006). Topics in herpesvirus genomics and evolution. Virus Res., 117, 90-104.
[17] Mettenleiter, T.C. (2002). Herpesvirus Assembly and Egress. J. Virol., 76(4), 1537-1547.
[18] Mossman, K., Sherburne, R., Lavery, C., Duncan, J. & Smiley, J. (2000). Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol., 74, 6287–6299.
[19] Quackenbush, S.L., Casey, R.N., Murcek, R.J., Paul, T.A., Work, T.M., Limpus, C.J., Chaves, A., duToit, L., Vasconcelos Perez, J., Aguirre, A., Spraker, T.R., Horrocks, J.A., Vermeer, L.A., Balazs, G.H. & Casey, J.W. (2001). Quantitative analysis of herpesvirus sequences from normal tissue and fibropapillomas of marine turtles with real time PCR. Virology, 287, 105– 111.
[20] Scott, E.S. & O’Hare, P. (2001). Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol., 75, 8818–8830.
[21] Shukla, D. & Spear, P.G. (2001). Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig., 108, 503–510.
[22] Spear, P.G., R.J. Eisenberg & Cohen, G.H.. (2000). Three classes of cell surface receptors for alphaherpesvirus entry. Virology, 275, 1–8.
[23] Spear, P.G. & Longnecker, R. (2003). Herpesvirus Entry: an Update. J. Virol., 77(19), 10179-10185.
[24] Wadsworth, S., Jacob, R.J. & Roizman, B. (1975). Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J. Virol., 15, 1487–1497.
[25] Weller, S.K. (2011). Alphaherpesviruses: Molecular Virology Preview. Caister Academic Press. Retrieved 9/6/10 from <http://www.horizonpress.com/alphaherpesviruses>.
[26] Williams, E.H.J., Bunkley-Williams, L., Peters, E.C., Pinto-Rodriguez, B., Matos-Morales, R., Mignucci-Giannoni, A.A., Hall, K.V., Rueda-Almonacid, J.V., Sybesma, J., Bonnelly de Calventi, I. & Boulon, R. (1994). An epizootic of cutaneous fibropapillomas in green turtles Chelonia mydas of the Caribbean: part of a panzootic? J. Aquat. Animal Health, 6, 70–78.
[27] Work, T.M., Balazs, G.H., Rameyer, R.A. & Morris, R.M. (2004). Retrospective pathology survey of green turtles (Chelonia mydas) with fibropapillomatosis in the Hawaiian Islands, 1993–2003. Dis. Aquat. Org., 62, 163-176.
Page authored for BIOL 375 Virology, September 2010
