Infectious Disease in the Neolithic: Difference between revisions
| Line 9: | Line 9: | ||
==Methods of Study== | ==Methods of Study== | ||
</b> | </b> | ||
The presence of infectious disease in the Neolithic can be examined in several ways. The most common is through the archaeological record. For decades, the presence of mass graves and skeletal markers of disease after excavation were the only available indicators of pathogens from thousands of years ago<ref name=Fuchs>[http://journals.sagepub.com/doi/10.1177/0959683619857230 Fuchs, Katharina, Christoph Rinne, Clara Drummer, Alexander Immel, Ben Krause-Kyora, and Almut Nebel. “Infectious Diseases and Neolithic Transformations: Evaluating Biological and Archaeological Proxies in the German Loess Zone between 5500 and 2500 BCE.” 2019. The Holocene 29 (10): 1545–57.]</ref>. As a result, this type of analysis has become very robust. The combination of various skeletal deformities and lesions can distinguish between similar diseases; tuberculosis and syphilis, for example, can both cause bone lesions, but syphilis does so specifically on the face and may affect the long bones like the tibia in unique ways<ref name=Vlok>[https://doi.org/10.5744/bi.2020.1000 Vlok, Melandri, Marc Oxenham, Kate Domett, Tran Thi Minh, Thi Mai Huong Nguyen, Hirofumi Matsumura, Hiep Hoang Trinh, et al. “Two Probable Cases of Infection with Treponema Pallidum during the Neolithic Period in Northern Vietnam (ca. 2000–1500 B.C.).” 2020. Bioarchaeology International 4 (1): 15–36.]</ref>. Unfortunately, these methods limit paleomicrobiology to the effects of the microbes and not the organisms themselves. As a result, skeletal paleopathology only identifies microbes that cause diseases that affect bone and have cases severe enough to be visible on the skeleton<ref name=Key>[https://doi.org/10.1038/s41559-020-1106-9 Key, Felix M., Cosimo Posth, Luis R. Esquivel-Gomez, Ron Hübler, Maria A. Spyrou, Gunnar U. Neumann, Anja Furtwängler, et al. “Emergence of Human-Adapted Salmonella Enterica Is Linked to the Neolithization Process.” 2020. Nature Ecology & Evolution 4 (3): 324–33.]</ref><ref name=Buikstra/>. Diseases caused by <i>Treponema</i> like syphilis and yaws, for example, only start permanently affecting bone when they reach the tertiary stage<ref name=Vlok/>. This stage only occurs in one of every three untreated cases, suggesting that diseases significantly affecting quality of life and population demographics may be seriously underrepresented<ref name=Vlok/>. | The presence of infectious disease in the Neolithic can be examined in several ways. The most common is through the archaeological record. For decades, the presence of mass graves and skeletal markers of disease after excavation were the only available indicators of pathogens from thousands of years ago<ref name=Fuchs>[http://journals.sagepub.com/doi/10.1177/0959683619857230 Fuchs, Katharina, Christoph Rinne, Clara Drummer, Alexander Immel, Ben Krause-Kyora, and Almut Nebel. “Infectious Diseases and Neolithic Transformations: Evaluating Biological and Archaeological Proxies in the German Loess Zone between 5500 and 2500 BCE.” 2019. The Holocene 29 (10): 1545–57.]</ref>. As a result, this type of analysis has become very robust. [[Image:M. tuberculosis.jpg|thumb|300px|left|Figure 2: Mycobacterium Tuberculosis Bacteria scanning electron micrograph Scanning electron micrograph of Mycobacterium tuberculosis bacteria. Photo Credit: Licensed under CC BY 2.0 DEED NIAID [https://www.flickr.com/photos/niaid/5149398656]]] The combination of various skeletal deformities and lesions can distinguish between similar diseases; tuberculosis and syphilis, for example, can both cause bone lesions, but syphilis does so specifically on the face and may affect the long bones like the tibia in unique ways<ref name=Vlok>[https://doi.org/10.5744/bi.2020.1000 Vlok, Melandri, Marc Oxenham, Kate Domett, Tran Thi Minh, Thi Mai Huong Nguyen, Hirofumi Matsumura, Hiep Hoang Trinh, et al. “Two Probable Cases of Infection with Treponema Pallidum during the Neolithic Period in Northern Vietnam (ca. 2000–1500 B.C.).” 2020. Bioarchaeology International 4 (1): 15–36.]</ref>. Unfortunately, these methods limit paleomicrobiology to the effects of the microbes and not the organisms themselves. As a result, skeletal paleopathology only identifies microbes that cause diseases that affect bone and have cases severe enough to be visible on the skeleton<ref name=Key>[https://doi.org/10.1038/s41559-020-1106-9 Key, Felix M., Cosimo Posth, Luis R. Esquivel-Gomez, Ron Hübler, Maria A. Spyrou, Gunnar U. Neumann, Anja Furtwängler, et al. “Emergence of Human-Adapted Salmonella Enterica Is Linked to the Neolithization Process.” 2020. Nature Ecology & Evolution 4 (3): 324–33.]</ref><ref name=Buikstra/>. Diseases caused by <i>Treponema</i> like syphilis and yaws, for example, only start permanently affecting bone when they reach the tertiary stage<ref name=Vlok/>. This stage only occurs in one of every three untreated cases, suggesting that diseases significantly affecting quality of life and population demographics may be seriously underrepresented<ref name=Vlok/>. | ||
In recent years, development of molecular methods like the polymerase chain reaction (PCR) and gene sequencing has resulted in powerful analysis by palaeomicrobiologists on the causative agents of diseases themselves<ref name=Donoghue>[https://doi.org/10.1002/9781119592112.ch34 Donoghue, Helen D. “Palaeomicrobiology of Human Infectious Diseases.” 2023. In Handbook of Archaeological Sciences, edited by A. Mark Pollard, Ruth Ann Armitage, and Cheryl A. Makarewicz, 1st ed., 677–96. Wiley.]</ref>. Furthermore, some microorganisms such as mycobacteria, to which tuberculosis and leprosy are related, can be detected by their very stable cell wall lipids <ref name=Donoghue/><ref name=Masson>[https://doi.org/10.1016/j.tube.2015.02.007 Masson, Muriel, Zsolt Bereczki, Erika Molnár, Helen D. Donoghue, David E. Minnikin, Oona Y.-C. Lee, Houdini H.T. Wu, Gurdyal S. Besra, Ian D. Bull, and György Pálfi. “7000 Year-Old Tuberculosis Cases from Hungary – Osteological and Biomolecular Evidence.” 2015. Tuberculosis 95 (June): S13–17.]</ref>. Ancient DNA (aDNA) from the Neolithic is often sampled from the environment or from bone itself <ref name=Bergfeldt/><ref name=Margaryan>[https://doi.org/10.1002/ece3.3924 Margaryan, Ashot, Henrik B. Hansen, Simon Rasmussen, Martin Sikora, Vyacheslav Moiseyev, Alexandr Khoklov, Andrey Epimakhov, et al. “Ancient Pathogen DNA in Human Teeth and Petrous Bones.” 2018. Ecology and Evolution 8 (6): 3534–42.]</ref>. With amplification, different species can be identified, characterized, and placed within a phylogenetic tree describing their evolutionary histories<ref name=Rascovan>[https://doi.org/10.1016/j.cell.2018.11.005 Rascovan, Nicolás, Karl-Göran Sjögren, Kristian Kristiansen, Rasmus Nielsen, Eske Willerslev, Christelle Desnues, and Simon Rasmussen. “Emergence and Spread of Basal Lineages of Yersinia Pestis during the Neolithic Decline.” 2019. Cell 176 (1–2): 295-305.e10.]</ref>. This has resulted in identification of pathogens previously unseen in the Neolithic; Pantovirus B19, <i>Yersenia pestis</i>, <i>Helocobacter pylori</i>, and hepatitis B, among others, have been found in Neolithic populations from aDNA alone<ref name=Fuchs/>. | In recent years, development of molecular methods like the polymerase chain reaction (PCR) and gene sequencing has resulted in powerful analysis by palaeomicrobiologists on the causative agents of diseases themselves<ref name=Donoghue>[https://doi.org/10.1002/9781119592112.ch34 Donoghue, Helen D. “Palaeomicrobiology of Human Infectious Diseases.” 2023. In Handbook of Archaeological Sciences, edited by A. Mark Pollard, Ruth Ann Armitage, and Cheryl A. Makarewicz, 1st ed., 677–96. Wiley.]</ref>. Furthermore, some microorganisms such as mycobacteria, to which tuberculosis and leprosy are related, can be detected by their very stable cell wall lipids <ref name=Donoghue/><ref name=Masson>[https://doi.org/10.1016/j.tube.2015.02.007 Masson, Muriel, Zsolt Bereczki, Erika Molnár, Helen D. Donoghue, David E. Minnikin, Oona Y.-C. Lee, Houdini H.T. Wu, Gurdyal S. Besra, Ian D. Bull, and György Pálfi. “7000 Year-Old Tuberculosis Cases from Hungary – Osteological and Biomolecular Evidence.” 2015. Tuberculosis 95 (June): S13–17.]</ref>. Ancient DNA (aDNA) from the Neolithic is often sampled from the environment or from bone itself <ref name=Bergfeldt/><ref name=Margaryan>[https://doi.org/10.1002/ece3.3924 Margaryan, Ashot, Henrik B. Hansen, Simon Rasmussen, Martin Sikora, Vyacheslav Moiseyev, Alexandr Khoklov, Andrey Epimakhov, et al. “Ancient Pathogen DNA in Human Teeth and Petrous Bones.” 2018. Ecology and Evolution 8 (6): 3534–42.]</ref>. With amplification, different species can be identified, characterized, and placed within a phylogenetic tree describing their evolutionary histories<ref name=Rascovan>[https://doi.org/10.1016/j.cell.2018.11.005 Rascovan, Nicolás, Karl-Göran Sjögren, Kristian Kristiansen, Rasmus Nielsen, Eske Willerslev, Christelle Desnues, and Simon Rasmussen. “Emergence and Spread of Basal Lineages of Yersinia Pestis during the Neolithic Decline.” 2019. Cell 176 (1–2): 295-305.e10.]</ref>. This has resulted in identification of pathogens previously unseen in the Neolithic; Pantovirus B19, <i>Yersenia pestis</i>, <i>Helocobacter pylori</i>, and hepatitis B, among others, have been found in Neolithic populations from aDNA alone<ref name=Fuchs/>. | ||
Revision as of 01:53, 15 April 2024
Introduction
The Neolithic, also known as the Stone Age, is a period between 10,000 and 2,200 BCE that is characterized by a shift from the hunter-gatherer lifestyle to one revolving around more permanent settlements and a newfound reliance on agriculture. With farming came, for the first time in human history, a consistent surplus of food. This allowed for rapid population growth and farming populations spread quickly throughout regions inhabited by hunter-gatherers[1]. Paleolithic people, who lived in the time period before the Neolithic, were constrained to relatively small, mobile populations in which the energetic cost of reproduction was quite high. It has been estimated that children would be raised only once every four years[2]. Farming and animal domestication allowed for the caloric needs of larger numbers of people to be met, and population size rapidly increased due to high fertility rates and migration patterns[3]. Altogether, this resulted in a large-scale shift towards larger, more concentrated communities, closer interactions with animals, and consistent use of the same landscapes for waste deposition and resource extraction.

These lifestyle changes create favorable conditions for the spread of disease into new niches. In fact, it is presumed that the Neolithic coincided with an increased impact of infectious disease on human populations. The emergence of human-adapted Salmonella strains, for example, is linked to the cultural transformations of the Neolithic, during which the sheer number of people made obligate parasitism a viable survival strategy[4]. Before the transition, small groups of hunter-gatherers in Africa consisted of only about 50 people and resulted in a low population density of about 30 people per square kilometer[2].The effective human population, therefore, was quite small, and any pathogens that specialized in human infection would have been relegated to a long period of stagnant growth. Ancestors of Mycobacterium tuberculosis would have only seen about 0.003% annual growth during the Pleistocene[2]. With larger populations, however, growth increased dramatically[2]. The effect of animal domestication in the spread of disease should not be underestimated. Evidence of systemic animal husbandry suggests a heavy reliance on animal labor and products throughout the Neolithic period, which presents greater opportunities for spillover events between species[5]. This is one of the leading theories for the evolution of human-specific tuberculosis[6]. While societies could now support rapidly increasing populations, the quality of life was not necessarily significantly improved. Elevated exposure to disease and higher workloads in more complex communities meant that individuals had high health risks[7]. Neolithic individuals also often present symptoms of nutritional and mineral deficiencies, which would increase disease risk[8][9].
Methods of Study
The presence of infectious disease in the Neolithic can be examined in several ways. The most common is through the archaeological record. For decades, the presence of mass graves and skeletal markers of disease after excavation were the only available indicators of pathogens from thousands of years ago[10]. As a result, this type of analysis has become very robust.
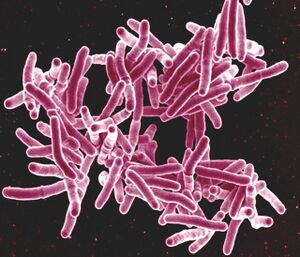
The combination of various skeletal deformities and lesions can distinguish between similar diseases; tuberculosis and syphilis, for example, can both cause bone lesions, but syphilis does so specifically on the face and may affect the long bones like the tibia in unique ways[8]. Unfortunately, these methods limit paleomicrobiology to the effects of the microbes and not the organisms themselves. As a result, skeletal paleopathology only identifies microbes that cause diseases that affect bone and have cases severe enough to be visible on the skeleton[4][11]. Diseases caused by Treponema like syphilis and yaws, for example, only start permanently affecting bone when they reach the tertiary stage[8]. This stage only occurs in one of every three untreated cases, suggesting that diseases significantly affecting quality of life and population demographics may be seriously underrepresented[8].
In recent years, development of molecular methods like the polymerase chain reaction (PCR) and gene sequencing has resulted in powerful analysis by palaeomicrobiologists on the causative agents of diseases themselves[12]. Furthermore, some microorganisms such as mycobacteria, to which tuberculosis and leprosy are related, can be detected by their very stable cell wall lipids [12][13]. Ancient DNA (aDNA) from the Neolithic is often sampled from the environment or from bone itself [14][15]. With amplification, different species can be identified, characterized, and placed within a phylogenetic tree describing their evolutionary histories[16]. This has resulted in identification of pathogens previously unseen in the Neolithic; Pantovirus B19, Yersenia pestis, Helocobacter pylori, and hepatitis B, among others, have been found in Neolithic populations from aDNA alone[10].
Major Pathogens
Recent study of infectious diseases in the Neolithic has revealed a variety of microbes affecting humans during the time period, especially those that are only detectable from DNA. These range from Pantovirus B19 to brucellosis, meningitis, and leprosy[10][17]. Listed here are several of the pathogens that have been extensively studied and are subjects of newly developing research.
Mycobacterium tuberculosis
Genetic evidence of Mycobacterium tuberculosis, the causative agent of tuberculosis, has been found as early as 5000 years ago [10]. It is most often identified as a Mycobacterium tuberculosis complex, a larger group that is recognized by standard DNA probes [18]The complex includes M. tuberculosis, M. bovis, M. africanum, and M. microti [18]. M. tuberculosis is one of the most common causes of tuberculosis, but M. bovis and M. africanum can result in similar symptoms in humans[18]. Of these, M. bovis mostly affects cattle but can infect humans if infected meat and dairy products are ingested, while M. africanum is responsible for the majority of tuberculosis cases in Africa [18]. M. microti specifically affects mice and voles[18].
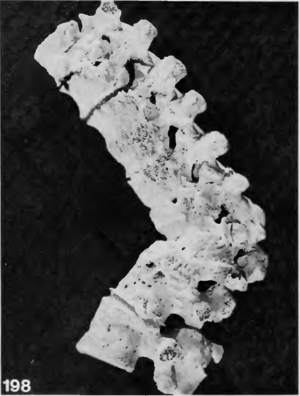
Cases of tuberculosis are often recorded in Neolithic burials. It is among the diseases most commonly reported in the archaeological record because it leaves diagnostic changes on human bone, often in the form of lesions and spinal collapse [10][11]. Calcifications in organs can also be indicative of previous tuberculosis infections [19].Individual burials in the Near East and Europe from early domestication phases in approximately 8800-7250 BCE are some of the earliest recorded cases of tuberculosis among humans [10]. In Europe, the earliest cases of skeletal tuberculosis date to about 5400-4800 BCE in Germany[10](Figure 3). While these demonstrate that tuberculosis was present within the Neolithic period, they also suggest that microbes within the complex were capable of infecting humans before the shift away from a hunter-gatherer lifestyle. One school of thought suggests that the bacteria evolved within Pleistocene megafauna before crossing over to humans. This is partially supported by evidence of ancient M. tuberculosis complex DNA found in a North American bison from 17,830 years ago[18][20]. Others, however, suggest that the complex emerged during the Neolithic, and not before. Recently, DNA extracted from a Swedish mummy showing signs of tuberculosis contributed to a molecular clock phylogeny that placed a common ancestor to the M. tuberculosis complex as late as 2000-6000 years before present[19]. If this is the case, it would suggest that the emergence of human tuberculosis was correlated with the Neolithic revolution.
Regardless of its origin before or during the Neolithic, tuberculosis and its associated complex were likely more active once people began practicing agriculture. In a model aiming to characterize the maintenance of tuberculosis over time, researchers reinforced the claim that tuberculosis growth rates were higher in the Neolithic (0.1%/year) than they had been previously (0.003%/year)[2] . This trend may be tied to worsening living conditions that correspond with a shift to a sedentary lifestyle.
Treponema
Treponematoses, describing several diseases caused by bacteria of the genus Treponema, have been identified in individuals from Northern Vietnam in 2000 BCE [8]. Treponematoses manifests in several commonly known diseases: yaws, from T. pallidum pertenue, pinta, from T. carateum, venerial syphilis, from T. pallidum pallidum, and endemic syphilis, from T. pallidum endemicum[8]. These diseases, like tuberculosis, can be surmised from the archaeological record because late stages can leave lesions in bone[8][11]. This method is particularly important because identification of ancient Treponema DNA is relatively lacking.
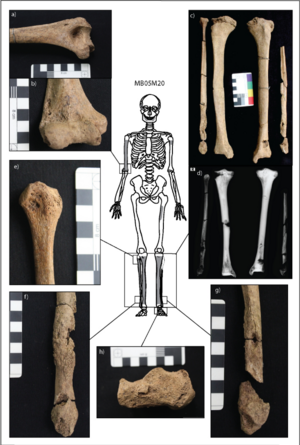
Soft, tumor-like growths and lesions called gumma can develop on the face and extracranial bone and are considered diagnostic for treponematoses[8]. Tibia deformation, called saber shin, and evidence of swelling in the digits point specifically to yaws in the two Vietnamese individuals[8]. Five additional individuals had symptoms suggesting treponematoses, but could not be confirmed as infected[8]. Because the individuals at the site were mostly juveniles, yaws may be the most likely candidate for infection at the site. Notably, the diagnostic lesions only occur in about 1% of patients with yaws, and they mostly occur in childhood[11]. Therefore, the presence of yaws in the archaeological record, and treponematoses as a whole, is likely underrespresentative of the actual presence of infection throughout the Neolithic (Figure 4).
Salmonella enterica
Evidence of Salmonella enterica affecting Neolithic populations has been found in several agrarian communities throughout Eurasia. Unlike M. tuberculosis and Treponema, Salmonella does not usually leave distinctive marks in human skeletons[4]. Instead, it is often identified via ancient DNA analysis. This method often relies on metagenomic samples from the environment. In the case of Salmonella, however, ancient microbial DNA is collected from teeth. Teeth are often highly preserved in the archaeological record and seem to preserve bacterial DNA better than other types of bone, with reports of high microbial diversity recorded from teeth[14][15]. Because teeth are vascularized – or contain blood vessels – in life, retrieval of microbial DNA from the teeth suggests that the microbe in question was present in high levels of an individual at the time of death[4]. In 2020, a study reported that eight Salmonella enterica genomes were collected from human teeth that were up to 6,500 years old[4]. With these data, it is clear that S. enterica in one form or another has been infecting human populations for thousands of years, well within the Neolithic. However, until 3,000 years ago, S. enterica strains may have been generalists, affecting a variety of mammals, until Neolithization made specifically infecting humans advantageous for some strains of the bacteria[4]. Similarity of ancient S. enterica genomes to those infecting pigs suggests some spillover or coevolution between strains infecting pigs and humans several thousand years ago[4].
Notably, S. enterica genomes have also been found in Bronze Age Crete with limited host adaptation to humans[21]. Presence of generalist strains after the Neolithic indicate that the Neolithic may not have affected S. enterica evolution as much as some other pathogens. While the bacteria could certainly infect humans, it may not have been advantageous to do so exclusively. That said, salmonellosis was likely still a serious concern for Neolithic people. DNA extraction from Stone Age Scandinavian farmers revealed S. entericain two individuals[14]. Because these two individuals were buried in the same grave, it is possible that salmonellosis was their cause of death[14]. Regardless, people infected with S. enterica could expect symptoms like stomach cramps, fever, and diarrhea, which would seriously reduce quality of life[14].
Yersinia pestis
The plague causing bacterium Yersinia pestis is most famous for the bubonic plague pandemic of the 1300s, but it had been infecting people for thousands of years before the famous Black Death pandemic. Based on ancient DNA analysis, Yersinia pestis has been identified in Scandinavian burials over 4,800 years old, the earliest known instance of plague to date[14]. A recent study from Germany's Kiel University has identified two new Y. pestis genomes from human remains at the Warburg necropolis, a site in Germany inhabited from 5,300-4,900 years before present[22]. This suggests a comparable or even earlier case of plague, but has yet to be peer-reviewed.
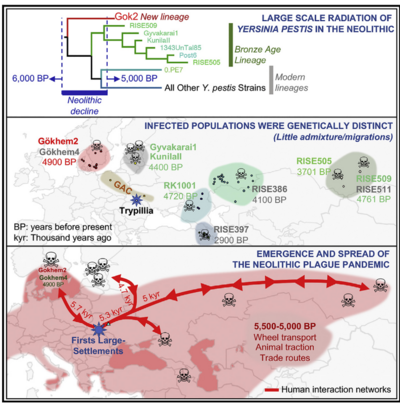
These cases represent individual infections by Y. pestis. Based on genomic analysis of a recently discovered ancient strain of Y. pestis, some have argued for a plague pandemic in Eurasia near the end of the Neolithic period[16]. Several different strains, like the newly discovered Gökheim 2, would have arisen in genetically distinct and relatively isolated population[16]. With increased trade networks that could be associated with the end of the Neolithic, these strains could have spread throughout the continent and contributed to the decline of prominent Neolithic societies [16]. During the end of the Neolithic, Y. pestis may have been capable of such extensive infection. For the most part, however, genomic characterization of Y. pestis strains present in the Neolithic, including Scandinavia where Gökheim 2 is presumed to have originated, suggests that they were not easily transmissible. In highly transmissible strains, the ymt gene allows for transmission via fleas[14]. In the earlier identified cases of Y. pestis infections, the mutation necessary for such transmission had not yet been acquired[14](Figure 5).
Parasites
In the Neolithic period, bacterial infections often occurred alongside parasitic infections by organisms like tapeworms and whipworms. These can be identified in the archaeological record without genomic analysis; eggs, most commonly of intestinal parasites, can be found within preserved soft tissue and coprolites[23][24][9]. Digital light microscopy on coprolites from a community near Stonehenge in 2500 BCE revealed a diverse array of parasite remnants, including helminth eggs, fish tapeworm (Dibothriocephalus dendriticus), and capillariid nematodes[24]. At a Neolithic site in Russia, coprolites similarly revealed evidence of parasites including the worms Diphyllobothrium and Dioctophyma renale and two trematodes including a river lancet fluke[23]. In both studies, the coprolites were mostly attributed to canids due to the diversity of parasites and the composition of bile[24][23]. At the Russian site, however, at least one human contributed to the sample[24]. This suggests not only that parasites may not have been incredibly host-specific, but that humans and dogs may have had a close enough relationship to spread parasites to one another. The close relationship between humans and canines has also been cited in the spread of bacteria. For example, dogs may have acted as a transient reservoir for Yersinia pestis[22].
A particularly interesting example of parasite infection in the Neolithic comes from the so-called "Iceman," otherwise known as Ötzi, an adult male mummy recovered from a glacier in 1991 and dated to 3350-3100 BCE[9]. Ötzi has been extensively studied over the last few decades, and research has revealed several chronic health conditions. One of these is an infection by the intestinal whipworm Trichuris trichiura, evidenced by many eggs found in Ötzi's intestines[9]. These parasites are ingested and result from poor sanitation and hygienic conditions[9]. Interestingly, Ötzi may have attempted to treat the symptoms from the T. trichiura by consuming the birch fungus Fomitopsis betulina, which was found on his person[9]. This fungus has anti-helminitic and anti-microbial properties, including substances that inhibit multi-drug resistance[25]. Ötzi's intestinal problems were likely intensified by a simultaneous infection by a particularly aggressive strain of Helicobacter pylori that, although often asymptomatic, likely caused active gastritis[9].
Critique of Expanding Disease in the Neolithic
The scale to which infectious diseases affected human populations in the Neolithic is also under debate. The orthodox field of thought emphasizes the unique conditions of the Neolithic that would promote large-scale changes in disease transmissibility and prevalence; large population sizes, sedentary lifestyles, and potential zoonotic spillover would all, in theory, contribute to drastic changes in the disease landscape. Some argue, however, that this is the result of overreliance on skeletal data and the assumption that a lack of visible skeletal anomalies equates to a healthy individual[26]. In fact, evidence of epidemics may exist in much older populations, Denisovans and Neanderthals, whose DNA is enriched in genetic pathways enhancing the immune response, particularly with T cells [27][26].
Tuberculosis presents an interesting case for this argument, with a dual adaptation to both high and low population densities. Tuberculosis has a high level of asymptomatic latency, which would allow reactivation at a later time, and has quite slow disease progression[6]. That said, its high virulence and transmission by aerosol suggest adaptation to a high density environment[6]. This suggests that the Mycobacterium tuberculosis complex, and presumably other pathogens, have historically adapted to infect hosts in both high and low density conditions.
Another critique of the infectious disease model of the Neolithic argues for the opposite – that epidemics and pandemics would not have occurred in the Neolithic, much less during earlier time periods. This is based on the composition of archaeological evidence for infection, in which aDNA and excavated burials provide clear evidence for single infections, but epidemics and pandemics are less supported[10]. A lack of consistent mass graves in Germany throughout the Neolithic support this conclusion, as epidemics with high mortality rates would be expected to coincide with a change in burial practices[10](Figure 6).
Conclusion
The Neolithic Period was a time of great change for human populations and the diseases that infected them. Research has historically been heavily based on the archaeological record and skeletal analysis from excavation. This has contributed to a robust understanding of how particular diseases like tuberculosis and treponematosis affect human bone, but resulted in the field focusing more on the effects of various diseases than their causative agents. Developments in genomic analysis, including PCR and genome sequencing of highly fragmentary ancient DNA, have revolutionized the field and provided evidence for a plethora of pathogenic microbes previously unrecognized in the Neolithic. These data have also contributed to the study of microbes that do cause recognizable anomalies in human remains, beginning to describe a complex evolutionary history. With rapid advances in the fields of paleopathology and paleomicrobiology, competing theories about the nature of infectious disease during the Neolithic revolution remain incredibly relevant.
References
- ↑ Chen, Xinfu, and Je-Chiang Tsai. “Spreading Speed in a Farmers and Hunter-Gatherers Model Arising from Neolithic Transition in Europe.” 2020. Journal de Mathématiques Pures et Appliquées 143 (November): 192–207.
- ↑ 2.0 2.1 2.2 2.3 2.4 Cardona, Pere-Joan, Martí Català, and Clara Prats. “The Origin and Maintenance of Tuberculosis Is Explained by the Induction of Smear-Negative Disease in the Paleolithic.” 2020. Pathogens 11 (3): 366
- ↑ Porčić, Marko, Tamara Blagojević, Jugoslav Pendić, and Sofija Stefanović. “The Neolithic Demographic Transition in the Central Balkans: Population Dynamics Reconstruction Based on New Radiocarbon Evidence.” 2021. Philosophical Transactions of the Royal Society B: Biological Sciences 376 (1816): 20190712.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Key, Felix M., Cosimo Posth, Luis R. Esquivel-Gomez, Ron Hübler, Maria A. Spyrou, Gunnar U. Neumann, Anja Furtwängler, et al. “Emergence of Human-Adapted Salmonella Enterica Is Linked to the Neolithization Process.” 2020. Nature Ecology & Evolution 4 (3): 324–33.
- ↑ Samartzidou, Eleni K. “Animal Disease Evidenced in the Bone Assemblage of a Late Neolithic Settlement in Greece: Implications for Animal Management.” 2024. International Journal of Paleopathology 44 (March): 126–39.
- ↑ 6.0 6.1 6.2 Buzic, Ileana, and Valentina Giuffra. “The Paleopathological Evidence on the Origins of Human Tuberculosis: A Review.” 2020. Journal of Preventive Medicine and Hygiene Vol. 61 No. 1s1 (April): E3 Pages.
- ↑ Larsen, Clark Spencer, Christopher J. Knüsel, Scott D. Haddow, Marin A. Pilloud, Marco Milella, Joshua W. Sadvari, Jessica Pearson, et al. “Bioarchaeology of Neolithic Çatalhöyük Reveals Fundamental Transitions in Health, Mobility, and Lifestyle in Early Farmers.” 2019. Proceedings of the National Academy of Sciences 116 (26): 12615–23.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 8.8 8.9 Vlok, Melandri, Marc Oxenham, Kate Domett, Tran Thi Minh, Thi Mai Huong Nguyen, Hirofumi Matsumura, Hiep Hoang Trinh, et al. “Two Probable Cases of Infection with Treponema Pallidum during the Neolithic Period in Northern Vietnam (ca. 2000–1500 B.C.).” 2020. Bioarchaeology International 4 (1): 15–36.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 Nerlich, Andreas G., Angelika Fleckinger, and Oliver Peschel. “Life and Diseases of the Neolithic Glacier Mummy ‘Ötzi.’” 2020. In The Handbook of Mummy Studies, edited by Dong Hoon Shin and Raffaella Bianucci, 1–22. Singapore: Springer Singapore.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 10.8 Fuchs, Katharina, Christoph Rinne, Clara Drummer, Alexander Immel, Ben Krause-Kyora, and Almut Nebel. “Infectious Diseases and Neolithic Transformations: Evaluating Biological and Archaeological Proxies in the German Loess Zone between 5500 and 2500 BCE.” 2019. The Holocene 29 (10): 1545–57.
- ↑ 11.0 11.1 11.2 11.3 Buikstra, J.E. ed. "Ortner's identification of pathological conditions in human skeletal remains." 2019.
- ↑ 12.0 12.1 Donoghue, Helen D. “Palaeomicrobiology of Human Infectious Diseases.” 2023. In Handbook of Archaeological Sciences, edited by A. Mark Pollard, Ruth Ann Armitage, and Cheryl A. Makarewicz, 1st ed., 677–96. Wiley.
- ↑ Masson, Muriel, Zsolt Bereczki, Erika Molnár, Helen D. Donoghue, David E. Minnikin, Oona Y.-C. Lee, Houdini H.T. Wu, Gurdyal S. Besra, Ian D. Bull, and György Pálfi. “7000 Year-Old Tuberculosis Cases from Hungary – Osteological and Biomolecular Evidence.” 2015. Tuberculosis 95 (June): S13–17.
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 14.7 Bergfeldt, Nora, Emrah Kırdök, Nikolay Oskolkov, Claudio Mirabello, Per Unneberg, Helena Malmström, Magdalena Fraser, et al. “Identification of Microbial Pathogens in Neolithic Scandinavian Humans.” 2024.Scientific Reports 14 (1): 5630.
- ↑ 15.0 15.1 Margaryan, Ashot, Henrik B. Hansen, Simon Rasmussen, Martin Sikora, Vyacheslav Moiseyev, Alexandr Khoklov, Andrey Epimakhov, et al. “Ancient Pathogen DNA in Human Teeth and Petrous Bones.” 2018. Ecology and Evolution 8 (6): 3534–42.
- ↑ 16.0 16.1 16.2 16.3 Rascovan, Nicolás, Karl-Göran Sjögren, Kristian Kristiansen, Rasmus Nielsen, Eske Willerslev, Christelle Desnues, and Simon Rasmussen. “Emergence and Spread of Basal Lineages of Yersinia Pestis during the Neolithic Decline.” 2019. Cell 176 (1–2): 295-305.e10.
- ↑ Henneberg, Maciej, Kara Holloway-Kew, and Teghan Lucas. “Human Major Infections: Tuberculosis, Treponematoses, Leprosy—A Paleopathological Perspective of Their Evolution.” 2021. Edited by David Caramelli. PLOS ONE 16 (2): e0243687.
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 Rothschild, Bruce M., Larry D. Martin, Galit Lev, Helen Bercovier, Gila Kahila Bar‐Gal, Charles Greenblatt, Helen Donoghue, Mark Spigelman, and David Brittain. “Mycobacterium Tuberculosis Complex DNA from an Extinct Bison Dated 17,000 Years before the Present.” 2001. Clinical Infectious Diseases 33 (3): 305–11.
- ↑ 19.0 19.1 Sabin et al. “A Seventeenth-Century Mycobacterium Tuberculosis Genome Supports a Neolithic Emergence of the Mycobacterium Tuberculosis Complex.” 2020. Genome Biology 21 (1): 201.
- ↑ Minnikin, David E, Oona Y-C Lee, Houdini Ht Wu, Gurdyal S Besra, and Helen D Donoghue. “Recognising the Broad Array of Approaches Available for the Diagnosis of Ancient Tuberculosis: Comment on ‘Infectious Diseases and Neolithic Transformations’ (Fuchs et al. 2019 The Holocene 29: 1545–1557).” 2020. The Holocene 30 (5): 781–83.
- ↑ Neumann, Gunnar U., Eirini Skourtanioti, Marta Burri, Elizabeth A. Nelson, Megan Michel, Alina N. Hiss, Photini J.P. McGeorge, et al. “Ancient Yersinia Pestis and Salmonella Enterica Genomes from Bronze Age Crete.” 2022. Current Biology 32 (16): 3641-3649.e8.
- ↑ 22.0 22.1 Krause-Kyora, Ben, Julian Susat, Magdalena Haller-Caskie, Joanna Bonczarowska, Nicolas Antonio Da Silva, Kerstin Schierhold, Michael Rind, et al. “Neolithic Humans and Dogs - Transient Reservoirs for Yersinia Pestis.” 2023.
- ↑ 23.0 23.1 23.2 Maicher, Celine, Yolaine Maigrot, Andrey Mazurkevich, Ekaterina Dolbunova, and Matthieu Le Bailly. “First Contribution of Paleoparasitology to the Study of Coprolites from the Neolithic Site Serteya II (NW Russia).” 2021. Journal of Archaeological Science: Reports 38 (August): 103093.
- ↑ 24.0 24.1 24.2 24.3 Mitchell, Piers D., Evilena Anastasiou, Helen L. Whelton, Ian D. Bull, Mike Parker Pearson, and Lisa-Marie Shillito. “Intestinal Parasites in the Neolithic Population Who Built Stonehenge (Durrington Walls, 2500 BCE).” 2022. Parasitology 149 (8): 1027–33.
- ↑ Pleszczyńska, Małgorzata, Marta K. Lemieszek, Marek Siwulski, Adrian Wiater, Wojciech Rzeski, and Janusz Szczodrak. “Fomitopsis Betulina (Formerly Piptoporus Betulinus): The Iceman’s Polypore Fungus with Modern Biotechnological Potential.” 2017. World Journal of Microbiology and Biotechnology 33 (5): 83.
- ↑ 26.0 26.1 Houldcroft, Charlotte J., and Simon Underdown. “Infectious Disease in the Pleistocene: Old Friends or Old Foes?” 2023. American Journal of Biological Anthropology 182 (4): 513–31.
- ↑ Vespasiani, Davide M., Guy S. Jacobs, Laura E. Cook, Nicolas Brucato, Matthew Leavesley, Christopher Kinipi, François-Xavier Ricaut, Murray P. Cox, and Irene Gallego Romero. “Denisovan Introgression Has Shaped the Immune System of Present-Day Papuans.” 2022. Edited by Anna Di Rienzo. PLOS Genetics 18 (12): e1010470.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024
