Korarchaeum cryptofilum: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
<i>Candidatus</i> Korarchaeum cryptofilum is a species of the proposed phylum <i>Korarchaeota</i>, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.<ref>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2449320/?tool=pmcentrez&report=abstract Nealson, Ken. “A Korarchaeote yields to genome sequencing.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105]</ref> The Archean phylum is found mainly in hydrothermal environments such as hot springs, shallow water, and deep ocean. The organism's genome has mainly been studied to open understanding of Archaean evolution due to its distant detachment on the Archaea phyla. | <i>Candidatus</i> Korarchaeum cryptofilum is a species of the proposed phylum <i>Korarchaeota</i>, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.<ref>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2449320/?tool=pmcentrez&report=abstract Nealson, Ken. “A Korarchaeote yields to genome sequencing.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105]</ref> The Archean phylum is found mainly in hydrothermal environments such as hot springs, shallow water, and deep ocean. The organism's genome has mainly been studied to open understanding of Archaean evolution due to its distant detachment on the Archaea phyla. | ||
[[Image:Korarchaeota.png|thumb|395px|right|<i>Ca</i>. K. cryptofilum. Notice rod shape of the Archaea. https://sites.google.com/site/chskorarchaeota1]] | |||
<br> | |||
==Discovery== | ==Discovery== | ||
The phylum <i>Korarchaeota</i> was only recently discovered in 1996 by a team studying Archaea in hot springs located in Yellowstone National Park. By sequencing the ribosomal RNA of many uncultivated archaeans found in a hot spring, two outlying sequences were found. The two outlying sequences were used as evidence for the proposal of a new phylum "Korarchaeota" | The phylum <i>Korarchaeota</i> was only recently discovered in 1996 by a team studying Archaea in hot springs located in Yellowstone National Park. By sequencing the ribosomal RNA of many uncultivated archaeans found in a hot spring, two outlying sequences were found. The two outlying sequences were used as evidence for the proposal of a new phylum "Korarchaeota"<ref name="Barns" /> | ||
= | |||
<br>At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of: | <br>At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of: | ||
| Line 23: | Line 20: | ||
<br><b>Subscript:</b> H<sub>2</sub>O | <br><b>Subscript:</b> H<sub>2</sub>O | ||
<br><b>Superscript:</b> Fe<sup>3+</sup> | <br><b>Superscript:</b> Fe<sup>3+</sup> | ||
<br> <br> | <br> <br> | ||
==Genetics and Structure== | ==Genetics and Structure== | ||
<br> <i>Korarchaeum cryptofilum</i> is | |||
<br> <i>Korarchaeum cryptofilum</i> is an ultrathin filamentous<ref name="Barns">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430366/?tool=pmcentrez&report=abstract Barns, S M et al. “Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences.” Proceedings of the National Academy of Sciences of the United States of America vol. 93,17 (1996): 9188-93. doi:10.1073/pnas.93.17.9188</ref> heterotroph, mostly living through anaerobic metabolism of peptide and amino acids. The organism uses enzymes such as peptidase, transaminases, and ATPase. <ref> [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2449320/?tool=pmcentrez&report=abstract Nealson, Ken. “A Korarchaeote yields to genome sequencing.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105]</ref> | |||
The genome of <i>Ca.</i> K. cryptofilum is 1,590,757 base pairs long with an average G+C content of 49%. The arCOG of the archaean phyla crenarchaeota and euryarchaeota when compared with the sequenced genome of <i>Ca.</i> K. cryptofilum, showed that the organism shares its replication, recombination, repair, and cell division genes with that of <i>crenarchaeota</i> while it shares most of its transcription and translation genes with <i>euryarcahaeota</i>. | The genome of <i>Ca.</i> K. cryptofilum is 1,590,757 base pairs long with an average G+C content of 49%. The arCOG of the archaean phyla crenarchaeota and euryarchaeota when compared with the sequenced genome of <i>Ca.</i> K. cryptofilum, showed that the organism shares its replication, recombination, repair, and cell division genes with that of <i>crenarchaeota</i> while it shares most of its transcription and translation genes with <i>euryarcahaeota</i>. | ||
<ref> [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3344838/> Miller-Coleman, Robin L et al. “Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning.” PloS one vol. 7,5 (2012): e35964. doi:10.1371/journal.pone.0035964]</ref> | <ref name=Miller-Coleman>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3344838/> Miller-Coleman, Robin L et al. “Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning.” PloS one vol. 7,5 (2012): e35964. doi:10.1371/journal.pone.0035964]</ref> | ||
<br> | <br> | ||
<i>Ca.</i> K. cryptofilum has three rRNA genes (16S, 23S, and 5S rRNA). The organism has shown to produce 33 23S r-proteins and 27 16S r-proteins on its rRNA operon. The species is also found to have 45 tRNA genes on its genome with one initiator and 45 elongator tRNAs.<ref | <i>Ca.</i> K. cryptofilum has three rRNA genes (16S, 23S, and 5S rRNA). The organism has shown to produce 33 23S r-proteins and 27 16S r-proteins on its rRNA operon. The species is also found to have 45 tRNA genes on its genome with one initiator and 45 elongator tRNAs.<ref name=Miller-Coleman/><ref>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430366/?Elkins, James G et al. “A korarchaeal genome reveals insights into the evolution of the Archaea.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,23 (2008): 8102-7. doi:10.1073/pnas.0801980105]</ref> | ||
| Line 57: | Line 53: | ||
<references /> | <references /> | ||
<br>Edited by [Charlie Stutz], student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol116/biol116_Fall_2013.html BIOL 116 Information in Living Systems], 2019, | <br>Edited by [Charlie Stutz], student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol116/biol116_Fall_2013.html BIOL 116 Information in Living Systems], 2019, | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | ||
Revision as of 00:53, 5 December 2019
Introduction
Candidatus Korarchaeum cryptofilum is a species of the proposed phylum Korarchaeota, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.[1] The Archean phylum is found mainly in hydrothermal environments such as hot springs, shallow water, and deep ocean. The organism's genome has mainly been studied to open understanding of Archaean evolution due to its distant detachment on the Archaea phyla.
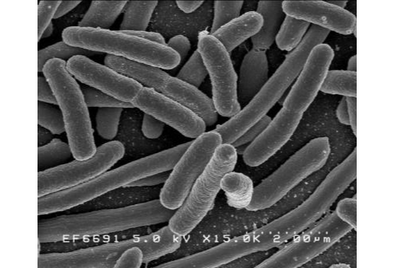
Discovery
The phylum Korarchaeota was only recently discovered in 1996 by a team studying Archaea in hot springs located in Yellowstone National Park. By sequencing the ribosomal RNA of many uncultivated archaeans found in a hot spring, two outlying sequences were found. The two outlying sequences were used as evidence for the proposal of a new phylum "Korarchaeota"[2]
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Genetics and Structure
Korarchaeum cryptofilum is an ultrathin filamentous[2] heterotroph, mostly living through anaerobic metabolism of peptide and amino acids. The organism uses enzymes such as peptidase, transaminases, and ATPase. [3]
The genome of Ca. K. cryptofilum is 1,590,757 base pairs long with an average G+C content of 49%. The arCOG of the archaean phyla crenarchaeota and euryarchaeota when compared with the sequenced genome of Ca. K. cryptofilum, showed that the organism shares its replication, recombination, repair, and cell division genes with that of crenarchaeota while it shares most of its transcription and translation genes with euryarcahaeota.
[4]
Ca. K. cryptofilum has three rRNA genes (16S, 23S, and 5S rRNA). The organism has shown to produce 33 23S r-proteins and 27 16S r-proteins on its rRNA operon. The species is also found to have 45 tRNA genes on its genome with one initiator and 45 elongator tRNAs.[4][5]
Information from Korarchaeum cryptofilum's genome reveals it is a
Microbiome
The Microbiome of
Include some current research, with a second image.
The population of the Korachaeota is generally minute in its microbiome living in the biome. However in some cases they constitute 7% of all Archaea.[6]
Conclusion
Overall text length should be at least 1,000 words (before counting references), with at least 2 images. Include at least 5 references under Reference section.
References
- ↑ Nealson, Ken. “A Korarchaeote yields to genome sequencing.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105
- ↑ 2.0 2.1 [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430366/?tool=pmcentrez&report=abstract Barns, S M et al. “Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences.” Proceedings of the National Academy of Sciences of the United States of America vol. 93,17 (1996): 9188-93. doi:10.1073/pnas.93.17.9188
- ↑ Nealson, Ken. “A Korarchaeote yields to genome sequencing.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105
- ↑ 4.0 4.1 > Miller-Coleman, Robin L et al. “Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning.” PloS one vol. 7,5 (2012): e35964. doi:10.1371/journal.pone.0035964
- ↑ James G et al. “A korarchaeal genome reveals insights into the evolution of the Archaea.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,23 (2008): 8102-7. doi:10.1073/pnas.0801980105
- ↑ Reigstad, Laila Johanne, et al. “Diversity and Abundance of Korarchaeota in Terrestrial Hot Springs of Iceland and Kamchatka.” ISME Journal: Multidisciplinary Journal of Microbial Ecology, vol. 4, no. 3, Mar. 2010, pp. 346–356. EBSCOhost, doi:10.1038/ismej.2009.126.
Edited by [Charlie Stutz], student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2019,
