Korarchaeum cryptofilum: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
<i>Candidatus</i> Korarchaeum cryptofilum is a species of the proposed phylum <i>Korarchaeota</i>, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.<ref name= "Nealson">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2449320/?tool=pmcentrez&report=abstract Nealson, Ken. | <i>Candidatus</i> Korarchaeum cryptofilum is a species of the proposed phylum <i>Korarchaeota</i>, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.<ref name= "Nealson">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2449320/?tool=pmcentrez&report=abstract Nealson, Ken. "A Korarchaeote yields to genome sequencing." Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105]</ref> However, there has been successful studies completed on the Archaeon's genome <ref name="Elkins" /> The Archaean phylum is found mainly in hydrothermal environments such as hot springs, shallow water, and deep ocean vents. The organism's genome has mainly been studied to open understanding of Archaean evolution due to its undetermined phylogeny placement for Archaea. The name of <i>Candidatus</i> Korarchaeum cryptofilum comes from the Greek words "<i>koros</i>" meaning "young man", "<i>kryptos</i>" meaning "hidden" in Greek, and "<i>filum</i>" which is Latin for "a thread". <ref name="Elkins" /> | ||
[[Image:Korarchaeota.png|thumb|395px|right|<i>Ca</i>. K. cryptofilum. Notice rod shape of the Archaea. https://sites.google.com/site/chskorarchaeota1]] | [[Image:Korarchaeota.png|thumb|395px|right|<i>Ca</i>. K. cryptofilum. Notice rod shape of the Archaea. https://sites.google.com/site/chskorarchaeota1]] | ||
==Discovery== | ==Discovery== | ||
The phylum <i>Korarchaeota</i> was only recently | The phylum <i>Korarchaeota</i> was only recently suggested in 1996 by a team of scientists studying Archaea in hot springs located in Yellowstone National Park. By sequencing the ribosomal RNA of many uncultivated Archaean's found in a hot spring, the phylum was discovered through two outlying archaean sequences found. The two outlying sequences were used as evidence for the proposal of a new phylum "Korarchaeota".<ref name="Barns" /> The organisms of the <i>Korarchaeota</i> phylum are found mainly in extremophile environments such as hot springs(such as <i>Candidatus</i> Korarchaeum cryptofilum) while others live in hot biomes like deep ocean floors and <ref name="Reigstad /> and on the coast thermal areas in Finland <ref name="Yan"> [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5787342/ Yan, Lijuan et al. "Distribution of Archaeal Communities along the Coast of the Gulf of Finland and Their Response to Oil Contamination." Frontiers in microbiology vol. 9 15. 23 Jan. 2018, doi:10.3389/fmicb.2018.00015]</ref> | ||
<i>Candidatus</i> Korarchaeum cryptofilum was most likely originally discovered when the phylum was discovered in 1996 and refered to as pJP27, however this is not confirmed. The archaeon was named in a published study in 2008, where a korarcaeota genome was sequenced using whole-genome shotgun sequencing.<ref name="Elkins" /> | <i>Candidatus</i> Korarchaeum cryptofilum was most likely originally discovered when the phylum was discovered in 1996 and refered to as pJP27, however this is not confirmed. The archaeon was named in a published study in 2008, where a korarcaeota genome was sequenced using whole-genome shotgun sequencing.<ref name="Elkins" /> | ||
<br> | |||
==Genetics and Structure== | ==Genetics and Structure== | ||
<i>Candidatus</i> Korarchaeum cryptofilum is an ultrathin filamentous<ref name="Barns">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC43212/ Barns, S M et al. "Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment." Proceedings of the National Academy of Sciences of the United States of America vol. 91,5 (1994): 1609-13. doi:10.1073/pnas.91.5.1609] </ref> heterotroph, mostly living through anaerobic metabolism of peptide fermentation and protein broken down into H<sub>2</sub>. The organism uses enzymes such as peptidase, transaminases, and ATPase. The organism has a partial citric acid cycle in its central metabolism. The enzymes in it, such as succinyl-CoA ligase are either used for the degradation or biosynthesis of the amino acid glutamate. <ref name="Nealson /> | |||
The genome of <i>Candidatus</i> Korarchaeum cryptofilum is a circular chromosome 1,590,757 base pairs long with an average G+C content of 49%. AUG is the start codon for 72.4% of the genome for understood protein-coding genes, with UUG and GUG at 17.6% and 10% respectively. Being a reference species for Archaean evolution, much of its genome is compared with organisms of other archaean phyla. The arCOG of <i>Crenarchaeota</i> and <i>Euryarchaeota</i> when compared with the sequenced genome of <i>Candidatus</i> Korarchaeum cryptofilum, showed that the organism shares similar replication, recombination, repair, and cell division genes with <i>Crenarchaeota</i> while it shares most of its transcription and translation genes with <i>Euryarcahaeota</i>. | |||
<ref name=Miller-Coleman>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3344838/> Miller-Coleman, Robin L et al. "Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning." PloS one vol. 7,5 (2012): e35964. doi:10.1371/journal.pone.0035964] </ref> The organism does not have the common Entner-Doudoroff pathways that are found in many <i>Crenarchaeota</i>.<ref name="Nealson" /> The unusual similarity to <i>Crenarchaeota</i> and <i>Euryarchaeota</i> in the archaeons genome suggest its genome has "retained features that represent an ancestral archaeal form, existing before the occurrence of the evolutionary bifurcation into <i>Crenarchaeota</i> and <i>Euryarchaeota</i>"<ref name="Herrick>Baltscheffsky, H. & Persson, B. J Mol Evol (2014) 78: 140</ref>. | |||
< | Chromosome replication is provoked by two <i>orc1/cdc6</i> homologous and one minichromosome maintenance protein complex. There are multiple DNA-dependent DNA polymerases. For transcription, <i>Candidatus</i> Korarchaeum cryptofilum has typical archaeal DNA-dependent RNA polymerase subunits. | ||
The Archaeon has three rRNA genes (16S, 23S, and 5S rRNA). The organism has shown to produce 33 23S r-proteins and 27 16S r-proteins on its rRNA operon. The species is also found to have 45 tRNA genes on its genome with one initiator and 45 elongator tRNAs.<ref name=Miller-Coleman/><ref name="Elkins">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430366/ Elkins, James G et al. "A korarchaeal genome reveals insights into the evolution of the Archaea." Proceedings of the National Academy of Sciences of the United States of America vol. 105,23 (2008): 8102-7. doi:10.1073/pnas.0801980105]</ref> | |||
<ref name= | <br><br> | ||
The size of <i>Candidatus</i> Korarchaeum cryptofilum is 0.16-0.18μm in diameter with a varying length. <ref Name="Elkins" />. | |||
<br><br> | |||
The phylogeny of the archaeon has been studied by looking at it large and small rRNA subunits. Conserved single-gene markers and conserved concatenated proteins. The lineage position of <i>Candidatus</i> Korarchaeum cryptofilum was proposed in three positions: the first argues the organism distantly branched from <i>Euryarchaeota</i>. The second and third argue the Archaeon branched off from <i>Crenarchaeota </i>. <ref name="Elkins" /> | |||
[[Image:Obsidian Pool.jpg|thumb|444px|right| Obsidian pool at Yellowstone National Park, similar to the habitat of <i>Candidatus</i> Korarchaeum cryptofilum https://ucmp.berkeley.edu/archaea/archaea.html]] | |||
<br> | <br> | ||
==Microbiome== | ==Microbiome== | ||
The Microbiome of <i>Candidatus</i> Korarchaeum cryptofilum is extreme compared to humans. The archaeon is found in obsidian pools at the Yellowstone National Park in Wyoming, United States. The overall phylum population of the <i> | The Microbiome of <i>Candidatus</i> Korarchaeum cryptofilum is extreme compared to humans. The archaeon is found in obsidian pools at the Yellowstone National Park in Wyoming, United States. The overall phylum population of the <i>Korarchaeota</i> is generally minute in its microbiome (with <i> Candidatus</i> Korarchaeum cryptofilum even less of that) living in the biome. However, in some cases they constitute 7% of all Archaea. The obsidian pools are very hot and high in saline concentration with only unicellular organisms living in it.<ref name="Reigstad">[http://web.b.ebscohost.com.libproxy.kenyon.edu/ehost/detail/detail?vid=0&sid=644f27fc-0fae-4646-8be1-1e8ac0adc257%40pdc-v-sessmgr05&bdata=JnNpdGU9ZWhvc3QtbGl2ZQ%3d%3d#AN=48092345&db=eih Reigstad, Laila Johanne, et al. "Diversity and Abundance of Korarchaeota in Terrestrial Hot Springs of Iceland and Kamchatka." ISME Journal: Multidisciplinary Journal of Microbial Ecology, vol. 4, no. 3, Mar. 2010, pp. 346-356. EBSCOhost, doi:10.1038/ismej.2009.126.]</ref> Obsidian pools at Yellow Stone National Park are full of thermophilic archaea who are able to withstand the acidic environment of some of the pools, more so than other microorganisms.<ref> [https://www.nps.gov/yell/learn/nature/thermophilic-archaea.htm "Thermophilic Archaea." National Parks Service, U.S. Department of the Interior]</ref> | ||
<br> | |||
==Conclusion== | |||
<i>Candidatus</i> Korarchaeum cryptofilum is a relative unknown Archaeon microorganism of the minor phylum <i>Korarchaeota</i>. Little research has been performed on it but key insights on Archaean evolution has been discovered, causing <i>Candidatus</i> Korarchaeum cryptofilum to be used as a reference species. The microorganism has never bene successfully cultivated which is the reason for the Archaeon to be considered a "Candidatus" species. What is known about the species has come through 16S ribosomal RNA.<ref name="Nealson" /> Studies on the Archaeon have raised many questions for the entire phylum of <i>Korarchaeota</i> including: "How did this apparently anciently diverging microbe evolve to be a heterotroph, dependent on amino acids, and devoid of biosynthetic abilities for many important things?" and " Is it a symbiont with an autotroph that supplies it with the "essentials" (amino acids, vitamins, purines, etc.) in exchange for hydrogen? Or is it just the first of many Korarchaeotes we will eventually encounter, leading to an appreciation for the entire group?"<ref name="Nealson" /> By successfully isolating the microorganism, much more can be understood about the organism and the fascinating phylum <i>Korarchaeota</i> as a whole. | |||
<br> | |||
=References= | =References= | ||
<references /> | <references /> | ||
Latest revision as of 01:30, 7 December 2019
Introduction
Candidatus Korarchaeum cryptofilum is a species of the proposed phylum Korarchaeota, or Xenarchaeota of the Archaea. The phylum is notoriously difficult to study given the environment in which it lives.[1] However, there has been successful studies completed on the Archaeon's genome [2] The Archaean phylum is found mainly in hydrothermal environments such as hot springs, shallow water, and deep ocean vents. The organism's genome has mainly been studied to open understanding of Archaean evolution due to its undetermined phylogeny placement for Archaea. The name of Candidatus Korarchaeum cryptofilum comes from the Greek words "koros" meaning "young man", "kryptos" meaning "hidden" in Greek, and "filum" which is Latin for "a thread". [2]
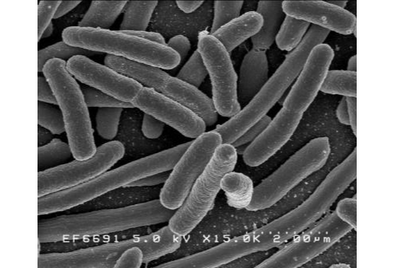
Discovery
The phylum Korarchaeota was only recently suggested in 1996 by a team of scientists studying Archaea in hot springs located in Yellowstone National Park. By sequencing the ribosomal RNA of many uncultivated Archaean's found in a hot spring, the phylum was discovered through two outlying archaean sequences found. The two outlying sequences were used as evidence for the proposal of a new phylum "Korarchaeota".[3] The organisms of the Korarchaeota phylum are found mainly in extremophile environments such as hot springs(such as Candidatus Korarchaeum cryptofilum) while others live in hot biomes like deep ocean floors and [4] and on the coast thermal areas in Finland [5]
Candidatus Korarchaeum cryptofilum was most likely originally discovered when the phylum was discovered in 1996 and refered to as pJP27, however this is not confirmed. The archaeon was named in a published study in 2008, where a korarcaeota genome was sequenced using whole-genome shotgun sequencing.[2]
Genetics and Structure
Candidatus Korarchaeum cryptofilum is an ultrathin filamentous[3] heterotroph, mostly living through anaerobic metabolism of peptide fermentation and protein broken down into H2. The organism uses enzymes such as peptidase, transaminases, and ATPase. The organism has a partial citric acid cycle in its central metabolism. The enzymes in it, such as succinyl-CoA ligase are either used for the degradation or biosynthesis of the amino acid glutamate. [1] The genome of Candidatus Korarchaeum cryptofilum is a circular chromosome 1,590,757 base pairs long with an average G+C content of 49%. AUG is the start codon for 72.4% of the genome for understood protein-coding genes, with UUG and GUG at 17.6% and 10% respectively. Being a reference species for Archaean evolution, much of its genome is compared with organisms of other archaean phyla. The arCOG of Crenarchaeota and Euryarchaeota when compared with the sequenced genome of Candidatus Korarchaeum cryptofilum, showed that the organism shares similar replication, recombination, repair, and cell division genes with Crenarchaeota while it shares most of its transcription and translation genes with Euryarcahaeota. [6] The organism does not have the common Entner-Doudoroff pathways that are found in many Crenarchaeota.[1] The unusual similarity to Crenarchaeota and Euryarchaeota in the archaeons genome suggest its genome has "retained features that represent an ancestral archaeal form, existing before the occurrence of the evolutionary bifurcation into Crenarchaeota and Euryarchaeota"[7].
Chromosome replication is provoked by two orc1/cdc6 homologous and one minichromosome maintenance protein complex. There are multiple DNA-dependent DNA polymerases. For transcription, Candidatus Korarchaeum cryptofilum has typical archaeal DNA-dependent RNA polymerase subunits.
The Archaeon has three rRNA genes (16S, 23S, and 5S rRNA). The organism has shown to produce 33 23S r-proteins and 27 16S r-proteins on its rRNA operon. The species is also found to have 45 tRNA genes on its genome with one initiator and 45 elongator tRNAs.[6][2]
The size of Candidatus Korarchaeum cryptofilum is 0.16-0.18μm in diameter with a varying length. [2].
The phylogeny of the archaeon has been studied by looking at it large and small rRNA subunits. Conserved single-gene markers and conserved concatenated proteins. The lineage position of Candidatus Korarchaeum cryptofilum was proposed in three positions: the first argues the organism distantly branched from Euryarchaeota. The second and third argue the Archaeon branched off from Crenarchaeota . [2]

Microbiome
The Microbiome of Candidatus Korarchaeum cryptofilum is extreme compared to humans. The archaeon is found in obsidian pools at the Yellowstone National Park in Wyoming, United States. The overall phylum population of the Korarchaeota is generally minute in its microbiome (with Candidatus Korarchaeum cryptofilum even less of that) living in the biome. However, in some cases they constitute 7% of all Archaea. The obsidian pools are very hot and high in saline concentration with only unicellular organisms living in it.[4] Obsidian pools at Yellow Stone National Park are full of thermophilic archaea who are able to withstand the acidic environment of some of the pools, more so than other microorganisms.[8]
Conclusion
Candidatus Korarchaeum cryptofilum is a relative unknown Archaeon microorganism of the minor phylum Korarchaeota. Little research has been performed on it but key insights on Archaean evolution has been discovered, causing Candidatus Korarchaeum cryptofilum to be used as a reference species. The microorganism has never bene successfully cultivated which is the reason for the Archaeon to be considered a "Candidatus" species. What is known about the species has come through 16S ribosomal RNA.[1] Studies on the Archaeon have raised many questions for the entire phylum of Korarchaeota including: "How did this apparently anciently diverging microbe evolve to be a heterotroph, dependent on amino acids, and devoid of biosynthetic abilities for many important things?" and " Is it a symbiont with an autotroph that supplies it with the "essentials" (amino acids, vitamins, purines, etc.) in exchange for hydrogen? Or is it just the first of many Korarchaeotes we will eventually encounter, leading to an appreciation for the entire group?"[1] By successfully isolating the microorganism, much more can be understood about the organism and the fascinating phylum Korarchaeota as a whole.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Nealson, Ken. "A Korarchaeote yields to genome sequencing." Proceedings of the National Academy of Sciences of the United States of America vol. 105,26 (2008): 8805-6. doi:10.1073/pnas.0804670105
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Elkins, James G et al. "A korarchaeal genome reveals insights into the evolution of the Archaea." Proceedings of the National Academy of Sciences of the United States of America vol. 105,23 (2008): 8102-7. doi:10.1073/pnas.0801980105
- ↑ 3.0 3.1 Barns, S M et al. "Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment." Proceedings of the National Academy of Sciences of the United States of America vol. 91,5 (1994): 1609-13. doi:10.1073/pnas.91.5.1609
- ↑ 4.0 4.1 Reigstad, Laila Johanne, et al. "Diversity and Abundance of Korarchaeota in Terrestrial Hot Springs of Iceland and Kamchatka." ISME Journal: Multidisciplinary Journal of Microbial Ecology, vol. 4, no. 3, Mar. 2010, pp. 346-356. EBSCOhost, doi:10.1038/ismej.2009.126.
- ↑ Yan, Lijuan et al. "Distribution of Archaeal Communities along the Coast of the Gulf of Finland and Their Response to Oil Contamination." Frontiers in microbiology vol. 9 15. 23 Jan. 2018, doi:10.3389/fmicb.2018.00015
- ↑ 6.0 6.1 > Miller-Coleman, Robin L et al. "Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning." PloS one vol. 7,5 (2012): e35964. doi:10.1371/journal.pone.0035964
- ↑ Baltscheffsky, H. & Persson, B. J Mol Evol (2014) 78: 140
- ↑ "Thermophilic Archaea." National Parks Service, U.S. Department of the Interior
Edited by [Charlie Stutz], student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2019,
