Magnetospirillum magnetotacticum: Difference between revisions
No edit summary |
|||
| (131 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image: | {{Uncurated}} | ||
[[Image:Marte179_microb_pic.jpg|frame|http://telem.openu.ac.il/courses/c20237/magneto.htm]] | |||
==Classification== | ==Classification== | ||
Bacteria (Kingdom); Proteobacteria (Phylum); Alphaproteobacteria (Class); Rhodospirillales (Order); Rhodospirillaceae (Class); Magnetospirillum (Family); Magnetospirillum (Genus) | Bacteria (Kingdom); Proteobacteria (Phylum); Alphaproteobacteria (Class); Rhodospirillales (Order); Rhodospirillaceae (Class); Magnetospirillum (Family); ''Magnetospirillum'' (Genus) [7] | ||
==Species== | ==Species== | ||
| Line 8: | Line 11: | ||
==Description and Significance== | ==Description and Significance== | ||
''Magnetospirillum magnetotacticum'' is a | ''Magnetospirillum magnetotacticum'' is a gram negative, helical, [http://en.wikipedia.org/wiki/Magnetotaxis magnetotactic], [http://en.wikipedia.org/wiki/Microaerophilic microaerophilic] spirillum. It can be isolated from areas of oxic to anoxic transition, typically associated with the sediment-water interface of many freshwater environments [1], [4]. First identified by Robert Blakemore in the 1980’s, this microbe was initially classified under the genus ''Aquaspirillum'' and given the name ''Aquaspirillum magnetotacticum''. It wasn't until 1992 that phylogenetic studies of the 16S rRNA sequence resulted in the creation of a new and distinct genus, ''Magnetospirillum'', under which ''A. magnetotacticum'' was reclassified [2]. It is a member of the [http://en.wikipedia.org/wiki/Magnetotactic_bacteria magnetotactic bacteria] (MTB) class, whose mobility is dependent upon the magnetic characteristics of the environment [6]. ''M. magnetotacticum'' remains one of the most important species of ''Magnetospirillum'' as it was the first to be discovered and is one of the few that is culturable [3]. To this day, it remains one of the best studied of the taxonomically and morphologically diverse MTB class [10]. This is a biologically, ecologically, and commercially relevant microbe that synthesizes high quality single-domain [http://en.wikipedia.org/wiki/Magnetite magnetite] crystals, far superior to those produced industrially. Some of its many potential uses including: exploring the evolutionary role of magnetism in higher organisms; the creation of models for [http://en.wikipedia.org/wiki/Biomineralization biomineralization]; applications as geobiological tracers; models for mineralization of sediment, [http://en.wikipedia.org/wiki/Bioremediation bioremediation]; contrast enhancers in MRI's; waste water treatments for removal of heavy metals and radionuclides; cell separation imaging; manufacturing of printing inks and magnetic tapes; and magnetic targeting of pharmaceuticals [4], [3]. Because many of the biosynthetic pathways employed by this microbe have yet to be discovered, it remains an active area of research. Plans to utilize the valuable magnetite from these organisms will require a better understanding of their various metabolic processes and nutritional requirements [3]. Current research is aimed at completing the genome sequence of ''M. magnetotacticum''. This will allow for further exploration of phylogenetic relationships and could lead to the identification of the magnetite biosynthetic genes [3]. | ||
==Genome Structure== | ==Genome Structure== | ||
''Magnetospirillum Magnetotacticum'' has a circular genome | Sequencing of the complete ''Magnetospirillum Magnetotacticum'' genome is still underway. Current findings indicated that the ''M. magnetotacticum'' has a single circular genome of 9,211,536 nucleotides, or about 4.3mb [13]. The genome contains large amounts of guanine and cytosine, 66%, a feature noted by Blakemore as a key difference between this microbe and those of the genus ''Aquaspririllum'' [7], [1]. The data regarding the protein coding regions in this genome vary among resources, however, the NCBI website indicates that 78% of the genome is coding [7]. There are no [http://en.wikipedia.org/wiki/Pseudogenes pseudogenes] present in the genome [7]. | ||
Though no extrachromosomal structures | Though no extrachromosomal structures were found when analyzing undigested ''M. magnetotacticum'' DNA, one study found possible evidence of a circular plasmid consisting of 40,000 base pairs, containing 16s rRNA, ''bra'' and ''por'' genes [13]. Previous studies have revealed that the ''por'' gene codes for an [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductase] that participates in reduction reactions [18]. It is probably involved in the reduction of iron during magnetite synthesis. These genes were linearized, allowing for their detection by enzyme digestion [13]. | ||
An | An unusual feature of the chromosome was identified by Weko and colleagues, they discovered a clustering of genes that contribute to this microbes magnetotaxicity [13]. The genes ''bfr'', which codes for the iron storage protein [http://en.wikipedia.org/wiki/Bacterioferritin bacterioferritin], and ''magA'', which codes for the magnetosomal membrane that surrounds magnetite crystals, were found to be located maximally in the same 17% of the genome [13]. The magnetosomal membrane is the basis for the finite characteristics of the magnetosome and plays a crucial role in aligning the magnetosomes in a biologically functional manner [17]. Another unique facet of this genome is the presence of two overlapping ''bfr'' genes. Visible as a base pair overlap, this set of ''bfr'' genes is of importance because very few magnetotactic bacteria code for two ''bfr'' genes. Of the bacteria that do, only the closely related magnetic strain AMB-1 possess this overlap [13]. This information could aid researchers during future phylogenetic inquiries regarding these microbes [13]. Regulation and expression could provide possible explanations for this gene overlap. Differential expression of the genes could vary the amounts of the bacterioferritin protein produced [13]. This would be an advantageous adaptation in habitats of varying iron availability [4], [10]. | ||
''Magnetospirillum | ''Magnetospirillum magnetotacticum'' was the first magnetotactic bacteria to be phylogenetically analyzed using 16s rRNA sequences, as it was the first magnetotactic bacteria (MTB) isolated in pure culture [4]. As more information is acquired about the genomic sequence of this microbe, the evolutionary role of its magnetic properties may be discovered and possibly exploited for biological, ecological, medical, pharmaceutical, and commercial purposes [3]. As a species specific mechanism for biomineralization is likely in effect, understanding the genes, enzymes, and mechanisms involved in [http://en.wikipedia.org/wiki/Biological_ice_nuclei bioprecipitation] of magnetite, would enable researchers to manipulate the size and shape of the magnetite crystals to suit their needs [12]. | ||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
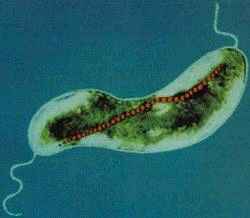
''M. magnetotacticum'' is a [http://en.wikipedia.org/wiki/Gram_negative gram negative], helical (clockwise), motile, spirilla that possesses two bipolar [http://en.wikipedia.org/wiki/Flagella flagella]. The organism ranges from 0.2-0.4 by 4.0-6.0µm and exhibits a wavelength of 1-2µm. ''M. magnetotacticum'' is a freshwater, [http://en.wikipedia.org/wiki/Denitrification denitrifying], [http://en.wikipedia.org/wiki/Chemoheterotroph chemoheterotroph][1], [10]. It can fix (N<sub>2</sub>) by reducing acetylene under microaerophilic conditions [10]. ''M. magnetotacticum'' is a well adapted microbe capable of utilizing a broad range of tricarboxylic acid cycle intermediates as both carbon and energy sources [1]. It is similarly able to select among co-oxidants: O<sub>2</sub> , NO<sub>3</sub> - , or Fe<sup>III</sup> as electron acceptors during metabolism [10]. How iron is acquired so efficiently by the organism from its iron poor environment remains an area of debate [4]. Previous studies suggesting that ''M. magnetotacticum'' produces a hydroxamate siderophore for iron acquisition, have been largely irreproducible [11], [4]. What is know, is that the microbe does readily acquire iron and that intracellular iron deposits make up approximately 2% of the cells total dry weight [11]. With the low levels of iron available in its habitat, it is speculated that ''M. magnetotacticum'' has highly specialized and efficient iron acquisition machinery [4]. It is currently believed that iron, Fe<sup>III</sup>, reduction may be coupled with iron acquisition, which is needed during intracellular magnetite (Fe<sub>3</sub>O<sub>4</sub>) crystal formation. The mechanism for magnetite biosynthesis is unknown [1], [3], [4]. Although, one proposed mechanism involves the reductive uptake of Fe<sup>III</sup> and subsequent reoxidiztion to a low density hydorous oxide, followed by dehydration to form the high density compound ferrihydrite [4]. The magnetite crystals synthesized by ''M. magnetotacticum'' are packaged into membrane enclosed compartments called [http://en.wikipedia.org/wiki/Magnetosome magnetosomes]. Magnetosomes have a cubooctahedral morphology and typically range in size from 40-50nm [1], [4]. They are organized into chains along the long (linear) axis of the cell and act cooperatively as a geomagnetic navigational apparatus [10], [4]. The magnetosomes are responsible for establishing a magnetic dipole in the organism that allow it to efficiently detect and migrate along the Earth's geomagnetic gradient [10]. The evolutionary advantages of magnetotaxis are not well understood. However, some speculate that magnetotaxis allows the microbe to traverse its environment in a more predictable and organized manner than microbes engaging in [http://en.wikipedia.org/wiki/Brownian_movement Brownian movement] [5]. It may also serve as a mechanism by which the bacteria can properly orient themselves in an optimal environment with reduced oxygen levels and high reduction potential [10]. Other proposed uses of intracellular iron uptake in ''M. magnetotacticum'' include: redox cycling, functions in iron homeostasis, and energy conservation [4]. As a member of the bacterial kingdom, ''M. magnetotacticum'' divides via [http://en.wikipedia.org/wiki/Binary_fission binary fission], in which one parent cell gives rise to two identical daughter cells. These bacteria generally form long chains of helical cells. In older cultures, under stressed conditions, coccoid bodies are abundant [1]. | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
'' | ''M. Magnetotacticum'' was first isolated by Robert Blakemore in the 1970's from Cedar Swamp, Massachusetts [1]. These bacteria can be isolated from oxic-anoxic transition zones consistent with the upper sediment layer of freshwater lakes, rivers, streams, ponds, bogs, marshes, and saturated soils [4]. ''M. magnetotacticum'' requires a reduced oxygen environment between 1-3% for survival, with optimal magnetite production occurring at oxygen partial pressures of 0.5-1.0 kPa [3], [10]. The peculiarity of the oxygen requirement for the survival of these organisms stems from their lack of key oxidative enzymes [1]. Studies done by Maratea and Blakemore, upon the initial discovery of the microbe, indicate that ''M. magnetotacticum'' does not possess either catalase or oxidase [1]. Some believe that because of this low oxygen requirement, the microbe may engage in [http://www.answers.com/topic/aerotaxis aerotaxis] as well as magnetotaxis [3]. It thrives at an optimal temperature of 30 degrees Celsius. And is extremely efficient at extracting iron even from iron poor habitats [4], [10]. ''M. magnetotacticum's'' shares an environment with a plethora of organisms, primarily other bacteria. In fact, bacteria contribute more to the overall biomass and biochemical activity than any other freshwater sedimentous organism [15]. Symbiotic relationships among microbes in this nutritionally stratified environment are crucial for the survival of this microbial community [15]. Organic and inorganic substrates as well as electron donors and acceptors are frequently exchanged among the various levels of sedimentation; typically the metabolic byproducts of one microbe serve as essential substrates for another within community [15]. ''M. magnetotacticum'' is optimally located above the anoxic zone inhabited by methanogens and sulfate reducing bacteria, which helps to alleviate some of the competitive pressure between the microbes for various nutrients [10]. Due to it's nutritional flexibility (it is capable of utilizing any of the tricarboxylic acid (TCA) cycle intermediates as both carbon and energy sources and several co-oxidants as electron acceptors), it is ideal to assume that these nutriental demands could easily be met by any number of microbes or higher organisms within the habitat. The release of TCA cycle intermediates occurs routinely by organisms engaging in glucose oxidation and during the fermentation of organic matter [19]. ''M. magnetotacticum'' plays an important role in [http://en.wikipedia.org/wiki/Biogeochemical_cycling biogeochemical cycles], having intimate involvement in the iron, carbon and nitrogen cycles [1], [4], [10]. As a denitrifying bacteria, it actively consumes and removes biologically available sources of nitrogen, acetylene, converting it to atmospheric nitrogen [10]. Likewise it skillfully removes iron from the environment during the production of the intracellular magnetite crystals thus, acting as a major component of the iron cycling system [1], [4], [10]. Carbon in the form of organic acids are utilized by the organism to provide energy and increase the microbes biomass [1]. The magnetite produced from these microbes is of considerable ecological importance [3], [4]. [http://en.wikipedia.org/wiki/Magnetofossils Magnetofossils] are among the most interesting footprints these organisms leave behind [3]. These are formed when an organism containing magnetite becomes fossilized. The magnetite within the fossil can be detected and plays an important role as a geobiological tracer [3], [16]. These fossilized remains are attributed to the magnetization of sediment [3]. ''M. magnetotacticum'' has no documented pathogenic activity. | ||
==References== | ==References== | ||
# Maratea, D., and Blakemore, R.P., 1981. ''Aquaspirillum magnetotacticum'' sp. nov., a Magnetic Spirillum. International Journal of Systematic Bacteriology. Vol 31.4:452-455. | |||
# Brenner,D. J., Bergey, D. H., Garrity, G. M., Krieg, N. R., <u>Bergey's manual of systematic bacteriology</u> Ed.2.Springer.2005. p:30-31. | |||
# "''Magnetospirillum magnetotacticum''" http://genome.jpi-psf.org/draft-microbes/magma/magma.home.html | |||
# Schülwer, D., 1999. Formation of Magnetosomes in Magnetotactic Bacteria. Journal of Molecular Microbiology and Biotechnology. 1(1):79-86 | |||
# Blakemore, R.P., and Frankel. 1981. Magnetite and magnetotaxis in microorganisms. Bioelectromagnetics. Vol 10.3:223-237 | |||
# "Magnetotactic bacteria" http://en.wikipedia.org/wiki/Magnetotactic_bacteria | |||
# "''Magnetosprillum magnetotacticum'' MS-1": http://www.uniprot.org/taxonomy/188 | |||
# "''Magnetosprillum magnetotacticum'' MS-1": http://genamics.com/cgi-bin/genamics/genomes/genomesearch.cgi?field=ID&query=955 | |||
# Eden, P., Schmidt, T., Blakemore, R., & Pace, N. 1991. Phylogenetic Analysis of ''Aquaspirillum magnetotacticum'' Using Polymerase Chain Reaction-Amplified 16S rRNA-Specific DNA. International Journal of Systematic Bacteriology. Vol 4.2:324-325. | |||
#Guerin, W., & Blakemore, R. 1992. Redox Cycling of Iron SUpports Growth and Magnetic Synthesis of ''Aquaspirillum magnetotacticum''. Applied & Envirnmental Microbiology. Vol 58.4:1102-1109 | |||
#Paoletti, L., & Blakemore, R. 1986. Hydroxamate production by ''Aquaspirillum magnetotacticum''. Journal of Bacteriology. Vol 167.1:73-76. | |||
# Ricci, J. C., Woodford, B. J., Kirschvink, J. L., Hoffman, M. R. 1991. Alteration of the Magnetic Properties of ''Aquaspirillum magnetotacticum'' by a Pulse Magnetization Technique. Applied and Environmental Microbioloy. Vol 57.1:3248-3254. | |||
#Bertaini, E., Weko, J., Phillips K., Gray. R., Kirschvink, J., 2001. Physical and Genetic Characterization of the Genome of "Magnetospirillum Magnetotacticum", strain MS-1. Gene. Vol. 264.2:257-263. | |||
#Blakemore, R., 1982. Magnetotactic Bacteria. Annual Reveiw of Microbiology. 1982.36:217-238. | |||
# Nealson, K.H. 1997. Sediment Bacteria: Who's There, What Are They Doing, and What's New? Annual Review of Earth and Planetary Sciences. Vol 25: 403-434 | |||
# "Magnetofossils" http://en.wikipedia.org/wiki/Magnetofossils | |||
# Lang ''et al''. 2006. Biogenic nanoparticles: production, characterization, and application of bacterial magnetososmes. Journal of Physics: Condensed Matter. Vol 38. | |||
# ''por'' genes: http://www.genecards.org/cgi-bin/carddisp.pl?gene=Por | |||
# Nelson, D. L., & Cox, M. M., 2008. Lehninger Principles of Biochemistry Fifth Ed.,W.H. Freeman and Co., 2008. Information collected from the "Glycolysis," "Citric Acid Cycle", and "Fermentation" chapters. | |||
====External Links:==== | |||
# Magnetotaxis: http://en.wikipedia.org/wiki/Magnetotaxis | |||
# Microaerophile: http://en.wikipedia.org/wiki/Microaerophilic | |||
# Magnetotactic Bacteria: http://en.wikipedia.org/wiki/Magnetotactic_bacteria | |||
# Magnetite: http://en.wikipedia.org/wiki/Magnetite | |||
# Bioremediation: http://en.wikipedia.org/wiki/Bioremediation | |||
# Biomineralisation: http://en.wikipedia.org/wiki/Biomineralisation | |||
# Pseudogenes: http://en.wikipedia.org/wiki/Pseudogenes | |||
# Oxidoreductase: http://en.wikipedia.org/wiki/Oxidoreductase | |||
# Bacterioferritin: http://en.wikipedia.org/wiki/Bacterioferritin | |||
# Bioprecipitation: http://en.wikipedia.org/wiki/Bioprecipitation | |||
# Gram Negative: http://en.wikipedia.org/wiki/Gram_negative | |||
# Flagella: http://en.wikipedia.org/wiki/Flagella | |||
# Denitrification: http://en.wikipedia.org/wiki/Denitrification | |||
# Chemoheterotroph: http://en.wikipedia.org/wiki/Chemoheterotroph | |||
# Magnetosomes: http://en.wikipedia.org/wiki/Magnetosome | |||
# Binary Fission: http://en.wikipedia.org/wiki/Binary_fission | |||
# Aerotaxis: http://www.answers.com/topic/aerotaxis | |||
# Biogeochemical cycles: http://en.wikipedia.org/wiki/Biogeochemical_cycles | |||
# Magnetofossils: http://en.wikipedia.org/wiki/Magnetofossils | |||
====Picture Link:==== | |||
# Magnetospirillum magnetotacticum: http://telem.openu.ac.il/courses/c20237/magneto.htm | |||
==Author== | ==Author== | ||
Page authored by Susan Jarosz and Megan Hull, students of [http://www.kbs.msu.edu/faculty/lennon/ Prof.Jay Lennon] at Michigan State University. | Page authored by Susan Jarosz and Megan Hull, students of [http://www.kbs.msu.edu/faculty/lennon/ Prof.Jay Lennon] at Michigan State University. | ||
Latest revision as of 18:59, 25 August 2010
Classification
Bacteria (Kingdom); Proteobacteria (Phylum); Alphaproteobacteria (Class); Rhodospirillales (Order); Rhodospirillaceae (Class); Magnetospirillum (Family); Magnetospirillum (Genus) [7]
Species
Magnetospirillum magnetotacticum (Synonym: Aquaspirillum magnetotacticum)
Description and Significance
Magnetospirillum magnetotacticum is a gram negative, helical, magnetotactic, microaerophilic spirillum. It can be isolated from areas of oxic to anoxic transition, typically associated with the sediment-water interface of many freshwater environments [1], [4]. First identified by Robert Blakemore in the 1980’s, this microbe was initially classified under the genus Aquaspirillum and given the name Aquaspirillum magnetotacticum. It wasn't until 1992 that phylogenetic studies of the 16S rRNA sequence resulted in the creation of a new and distinct genus, Magnetospirillum, under which A. magnetotacticum was reclassified [2]. It is a member of the magnetotactic bacteria (MTB) class, whose mobility is dependent upon the magnetic characteristics of the environment [6]. M. magnetotacticum remains one of the most important species of Magnetospirillum as it was the first to be discovered and is one of the few that is culturable [3]. To this day, it remains one of the best studied of the taxonomically and morphologically diverse MTB class [10]. This is a biologically, ecologically, and commercially relevant microbe that synthesizes high quality single-domain magnetite crystals, far superior to those produced industrially. Some of its many potential uses including: exploring the evolutionary role of magnetism in higher organisms; the creation of models for biomineralization; applications as geobiological tracers; models for mineralization of sediment, bioremediation; contrast enhancers in MRI's; waste water treatments for removal of heavy metals and radionuclides; cell separation imaging; manufacturing of printing inks and magnetic tapes; and magnetic targeting of pharmaceuticals [4], [3]. Because many of the biosynthetic pathways employed by this microbe have yet to be discovered, it remains an active area of research. Plans to utilize the valuable magnetite from these organisms will require a better understanding of their various metabolic processes and nutritional requirements [3]. Current research is aimed at completing the genome sequence of M. magnetotacticum. This will allow for further exploration of phylogenetic relationships and could lead to the identification of the magnetite biosynthetic genes [3].
Genome Structure
Sequencing of the complete Magnetospirillum Magnetotacticum genome is still underway. Current findings indicated that the M. magnetotacticum has a single circular genome of 9,211,536 nucleotides, or about 4.3mb [13]. The genome contains large amounts of guanine and cytosine, 66%, a feature noted by Blakemore as a key difference between this microbe and those of the genus Aquaspririllum [7], [1]. The data regarding the protein coding regions in this genome vary among resources, however, the NCBI website indicates that 78% of the genome is coding [7]. There are no pseudogenes present in the genome [7]. Though no extrachromosomal structures were found when analyzing undigested M. magnetotacticum DNA, one study found possible evidence of a circular plasmid consisting of 40,000 base pairs, containing 16s rRNA, bra and por genes [13]. Previous studies have revealed that the por gene codes for an oxidoreductase that participates in reduction reactions [18]. It is probably involved in the reduction of iron during magnetite synthesis. These genes were linearized, allowing for their detection by enzyme digestion [13]. An unusual feature of the chromosome was identified by Weko and colleagues, they discovered a clustering of genes that contribute to this microbes magnetotaxicity [13]. The genes bfr, which codes for the iron storage protein bacterioferritin, and magA, which codes for the magnetosomal membrane that surrounds magnetite crystals, were found to be located maximally in the same 17% of the genome [13]. The magnetosomal membrane is the basis for the finite characteristics of the magnetosome and plays a crucial role in aligning the magnetosomes in a biologically functional manner [17]. Another unique facet of this genome is the presence of two overlapping bfr genes. Visible as a base pair overlap, this set of bfr genes is of importance because very few magnetotactic bacteria code for two bfr genes. Of the bacteria that do, only the closely related magnetic strain AMB-1 possess this overlap [13]. This information could aid researchers during future phylogenetic inquiries regarding these microbes [13]. Regulation and expression could provide possible explanations for this gene overlap. Differential expression of the genes could vary the amounts of the bacterioferritin protein produced [13]. This would be an advantageous adaptation in habitats of varying iron availability [4], [10]. Magnetospirillum magnetotacticum was the first magnetotactic bacteria to be phylogenetically analyzed using 16s rRNA sequences, as it was the first magnetotactic bacteria (MTB) isolated in pure culture [4]. As more information is acquired about the genomic sequence of this microbe, the evolutionary role of its magnetic properties may be discovered and possibly exploited for biological, ecological, medical, pharmaceutical, and commercial purposes [3]. As a species specific mechanism for biomineralization is likely in effect, understanding the genes, enzymes, and mechanisms involved in bioprecipitation of magnetite, would enable researchers to manipulate the size and shape of the magnetite crystals to suit their needs [12].
Cell Structure, Metabolism and Life Cycle
M. magnetotacticum is a gram negative, helical (clockwise), motile, spirilla that possesses two bipolar flagella. The organism ranges from 0.2-0.4 by 4.0-6.0µm and exhibits a wavelength of 1-2µm. M. magnetotacticum is a freshwater, denitrifying, chemoheterotroph[1], [10]. It can fix (N2) by reducing acetylene under microaerophilic conditions [10]. M. magnetotacticum is a well adapted microbe capable of utilizing a broad range of tricarboxylic acid cycle intermediates as both carbon and energy sources [1]. It is similarly able to select among co-oxidants: O2 , NO3 - , or FeIII as electron acceptors during metabolism [10]. How iron is acquired so efficiently by the organism from its iron poor environment remains an area of debate [4]. Previous studies suggesting that M. magnetotacticum produces a hydroxamate siderophore for iron acquisition, have been largely irreproducible [11], [4]. What is know, is that the microbe does readily acquire iron and that intracellular iron deposits make up approximately 2% of the cells total dry weight [11]. With the low levels of iron available in its habitat, it is speculated that M. magnetotacticum has highly specialized and efficient iron acquisition machinery [4]. It is currently believed that iron, FeIII, reduction may be coupled with iron acquisition, which is needed during intracellular magnetite (Fe3O4) crystal formation. The mechanism for magnetite biosynthesis is unknown [1], [3], [4]. Although, one proposed mechanism involves the reductive uptake of FeIII and subsequent reoxidiztion to a low density hydorous oxide, followed by dehydration to form the high density compound ferrihydrite [4]. The magnetite crystals synthesized by M. magnetotacticum are packaged into membrane enclosed compartments called magnetosomes. Magnetosomes have a cubooctahedral morphology and typically range in size from 40-50nm [1], [4]. They are organized into chains along the long (linear) axis of the cell and act cooperatively as a geomagnetic navigational apparatus [10], [4]. The magnetosomes are responsible for establishing a magnetic dipole in the organism that allow it to efficiently detect and migrate along the Earth's geomagnetic gradient [10]. The evolutionary advantages of magnetotaxis are not well understood. However, some speculate that magnetotaxis allows the microbe to traverse its environment in a more predictable and organized manner than microbes engaging in Brownian movement [5]. It may also serve as a mechanism by which the bacteria can properly orient themselves in an optimal environment with reduced oxygen levels and high reduction potential [10]. Other proposed uses of intracellular iron uptake in M. magnetotacticum include: redox cycling, functions in iron homeostasis, and energy conservation [4]. As a member of the bacterial kingdom, M. magnetotacticum divides via binary fission, in which one parent cell gives rise to two identical daughter cells. These bacteria generally form long chains of helical cells. In older cultures, under stressed conditions, coccoid bodies are abundant [1].
Ecology and Pathogenesis
M. Magnetotacticum was first isolated by Robert Blakemore in the 1970's from Cedar Swamp, Massachusetts [1]. These bacteria can be isolated from oxic-anoxic transition zones consistent with the upper sediment layer of freshwater lakes, rivers, streams, ponds, bogs, marshes, and saturated soils [4]. M. magnetotacticum requires a reduced oxygen environment between 1-3% for survival, with optimal magnetite production occurring at oxygen partial pressures of 0.5-1.0 kPa [3], [10]. The peculiarity of the oxygen requirement for the survival of these organisms stems from their lack of key oxidative enzymes [1]. Studies done by Maratea and Blakemore, upon the initial discovery of the microbe, indicate that M. magnetotacticum does not possess either catalase or oxidase [1]. Some believe that because of this low oxygen requirement, the microbe may engage in aerotaxis as well as magnetotaxis [3]. It thrives at an optimal temperature of 30 degrees Celsius. And is extremely efficient at extracting iron even from iron poor habitats [4], [10]. M. magnetotacticum's shares an environment with a plethora of organisms, primarily other bacteria. In fact, bacteria contribute more to the overall biomass and biochemical activity than any other freshwater sedimentous organism [15]. Symbiotic relationships among microbes in this nutritionally stratified environment are crucial for the survival of this microbial community [15]. Organic and inorganic substrates as well as electron donors and acceptors are frequently exchanged among the various levels of sedimentation; typically the metabolic byproducts of one microbe serve as essential substrates for another within community [15]. M. magnetotacticum is optimally located above the anoxic zone inhabited by methanogens and sulfate reducing bacteria, which helps to alleviate some of the competitive pressure between the microbes for various nutrients [10]. Due to it's nutritional flexibility (it is capable of utilizing any of the tricarboxylic acid (TCA) cycle intermediates as both carbon and energy sources and several co-oxidants as electron acceptors), it is ideal to assume that these nutriental demands could easily be met by any number of microbes or higher organisms within the habitat. The release of TCA cycle intermediates occurs routinely by organisms engaging in glucose oxidation and during the fermentation of organic matter [19]. M. magnetotacticum plays an important role in biogeochemical cycles, having intimate involvement in the iron, carbon and nitrogen cycles [1], [4], [10]. As a denitrifying bacteria, it actively consumes and removes biologically available sources of nitrogen, acetylene, converting it to atmospheric nitrogen [10]. Likewise it skillfully removes iron from the environment during the production of the intracellular magnetite crystals thus, acting as a major component of the iron cycling system [1], [4], [10]. Carbon in the form of organic acids are utilized by the organism to provide energy and increase the microbes biomass [1]. The magnetite produced from these microbes is of considerable ecological importance [3], [4]. Magnetofossils are among the most interesting footprints these organisms leave behind [3]. These are formed when an organism containing magnetite becomes fossilized. The magnetite within the fossil can be detected and plays an important role as a geobiological tracer [3], [16]. These fossilized remains are attributed to the magnetization of sediment [3]. M. magnetotacticum has no documented pathogenic activity.
References
- Maratea, D., and Blakemore, R.P., 1981. Aquaspirillum magnetotacticum sp. nov., a Magnetic Spirillum. International Journal of Systematic Bacteriology. Vol 31.4:452-455.
- Brenner,D. J., Bergey, D. H., Garrity, G. M., Krieg, N. R., Bergey's manual of systematic bacteriology Ed.2.Springer.2005. p:30-31.
- "Magnetospirillum magnetotacticum" http://genome.jpi-psf.org/draft-microbes/magma/magma.home.html
- Schülwer, D., 1999. Formation of Magnetosomes in Magnetotactic Bacteria. Journal of Molecular Microbiology and Biotechnology. 1(1):79-86
- Blakemore, R.P., and Frankel. 1981. Magnetite and magnetotaxis in microorganisms. Bioelectromagnetics. Vol 10.3:223-237
- "Magnetotactic bacteria" http://en.wikipedia.org/wiki/Magnetotactic_bacteria
- "Magnetosprillum magnetotacticum MS-1": http://www.uniprot.org/taxonomy/188
- "Magnetosprillum magnetotacticum MS-1": http://genamics.com/cgi-bin/genamics/genomes/genomesearch.cgi?field=ID&query=955
- Eden, P., Schmidt, T., Blakemore, R., & Pace, N. 1991. Phylogenetic Analysis of Aquaspirillum magnetotacticum Using Polymerase Chain Reaction-Amplified 16S rRNA-Specific DNA. International Journal of Systematic Bacteriology. Vol 4.2:324-325.
- Guerin, W., & Blakemore, R. 1992. Redox Cycling of Iron SUpports Growth and Magnetic Synthesis of Aquaspirillum magnetotacticum. Applied & Envirnmental Microbiology. Vol 58.4:1102-1109
- Paoletti, L., & Blakemore, R. 1986. Hydroxamate production by Aquaspirillum magnetotacticum. Journal of Bacteriology. Vol 167.1:73-76.
- Ricci, J. C., Woodford, B. J., Kirschvink, J. L., Hoffman, M. R. 1991. Alteration of the Magnetic Properties of Aquaspirillum magnetotacticum by a Pulse Magnetization Technique. Applied and Environmental Microbioloy. Vol 57.1:3248-3254.
- Bertaini, E., Weko, J., Phillips K., Gray. R., Kirschvink, J., 2001. Physical and Genetic Characterization of the Genome of "Magnetospirillum Magnetotacticum", strain MS-1. Gene. Vol. 264.2:257-263.
- Blakemore, R., 1982. Magnetotactic Bacteria. Annual Reveiw of Microbiology. 1982.36:217-238.
- Nealson, K.H. 1997. Sediment Bacteria: Who's There, What Are They Doing, and What's New? Annual Review of Earth and Planetary Sciences. Vol 25: 403-434
- "Magnetofossils" http://en.wikipedia.org/wiki/Magnetofossils
- Lang et al. 2006. Biogenic nanoparticles: production, characterization, and application of bacterial magnetososmes. Journal of Physics: Condensed Matter. Vol 38.
- por genes: http://www.genecards.org/cgi-bin/carddisp.pl?gene=Por
- Nelson, D. L., & Cox, M. M., 2008. Lehninger Principles of Biochemistry Fifth Ed.,W.H. Freeman and Co., 2008. Information collected from the "Glycolysis," "Citric Acid Cycle", and "Fermentation" chapters.
External Links:
- Magnetotaxis: http://en.wikipedia.org/wiki/Magnetotaxis
- Microaerophile: http://en.wikipedia.org/wiki/Microaerophilic
- Magnetotactic Bacteria: http://en.wikipedia.org/wiki/Magnetotactic_bacteria
- Magnetite: http://en.wikipedia.org/wiki/Magnetite
- Bioremediation: http://en.wikipedia.org/wiki/Bioremediation
- Biomineralisation: http://en.wikipedia.org/wiki/Biomineralisation
- Pseudogenes: http://en.wikipedia.org/wiki/Pseudogenes
- Oxidoreductase: http://en.wikipedia.org/wiki/Oxidoreductase
- Bacterioferritin: http://en.wikipedia.org/wiki/Bacterioferritin
- Bioprecipitation: http://en.wikipedia.org/wiki/Bioprecipitation
- Gram Negative: http://en.wikipedia.org/wiki/Gram_negative
- Flagella: http://en.wikipedia.org/wiki/Flagella
- Denitrification: http://en.wikipedia.org/wiki/Denitrification
- Chemoheterotroph: http://en.wikipedia.org/wiki/Chemoheterotroph
- Magnetosomes: http://en.wikipedia.org/wiki/Magnetosome
- Binary Fission: http://en.wikipedia.org/wiki/Binary_fission
- Aerotaxis: http://www.answers.com/topic/aerotaxis
- Biogeochemical cycles: http://en.wikipedia.org/wiki/Biogeochemical_cycles
- Magnetofossils: http://en.wikipedia.org/wiki/Magnetofossils
Picture Link:
- Magnetospirillum magnetotacticum: http://telem.openu.ac.il/courses/c20237/magneto.htm
Author
Page authored by Susan Jarosz and Megan Hull, students of Prof.Jay Lennon at Michigan State University.