Marinomonas artica: Difference between revisions
No edit summary |
|||
| Line 9: | Line 9: | ||
Bacteria; Pseudomonadota; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; | Bacteria; Pseudomonadota; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Marinomonas | ||
Revision as of 17:53, 12 December 2022
Classification
Bacteria; Pseudomonadota; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Marinomonas
Species
|
NCBI: [1] |
Marinomonas artica
Description and Significance
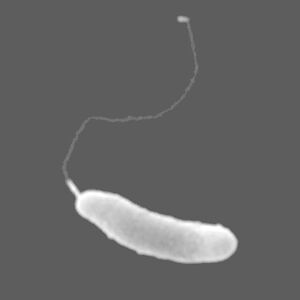
Marinomonas artica is a gram-negative, rod-shaped, and novel psychrotolerant bacteria that was first isolated from sea-ice just off the Canadian Basin of the Artic Ocean.
Psychrotolerant organisms like Marinomonas artica, are able to thrive in cold environments, degrading organic material and releasing necessary nutrients into the environment. They are also a great model organism to study to further our knowledge about psychrotolerant organisms, and just how they operate and survive in these harsh conditions.
Genome Structure
Not much genome sequence or research has been done on Marinomonas artica. However, just like other microorganisms, M. artica has one chromosome, yet no defined shape is cited for it. We can infer a possible circular chromosome through phylogeny, as its relative Marinomonas posidonica has been determined to have a circular chromosome as opposed to linear (Lucas-Elío et al., 2012; Chunming et al. 2014). M. artica is 4.5 Mb in length (ungapped) and 44.5% of its content is guanine-cytosine (NIH, 2020; Zhang et al., 2008).
The phospholipids and other fatty acids M. artica's genome codes for are phosphatidylethanolamine, phosphatidylglycerol, C10 : 0 3-OH (31.78 %), C18 : 1 ω7c (27.50 %), and iso-C15 : 0 2-OH and/or C16 : 1 ω7c (19.22 %). It was through phylogenetic analysis of the 16S rRNA gene sequence that the strain was labeled 328T and determined to be of the genius Marinomonas. (Zhang et al., 2008).
Cell Structure, Metabolism and Life Cycle
While the research of the Marinomonas genus and M. artica in particular is not vast, we do know a few key components of its structure along with how it metabolizes and thrives in its environment.
Marinomonas artica, along with many others of its Marinomonas genus, is a curved-rod-shaped bacterial cell with a flagellum that allows it to move about its icy and marine environment. It is about 0.3–0.4×1.2–2.6 μm in size and grows aerobically and produces catalase and cytochrome oxidase as a byproduct. Another component that protects M. artica and provides tolerance to the harsh sea ice it lives in is the major phospholipids, phosphatidylethanolamine and phosphatidylglycerol, it possesses. These phospholipids, along with many other fatty acids, regulate transportation in the cell and the processing of proteins (Zhang et al., 2008).
In contrast to its phylogenetic relatives, M. artica only uses a variety of carbon compounds as its sole source of energy. Though carbon compounds are M. artica's only energy source, the range of compounds are extremely broad and are named as followed: glucose, mannitol, d-mannose, sucrose, d-sorbitol, d-arabinose, l-rhamnose, maltose, fructose, d-galactose, erythritol, melibiose, glycerol, cellobiose, raffinose, xylose, melezitose, trehalose, l-proline, l-alanine, l-histidine, gluconate, malate and phenylacetic acid (Zhang et al., 2008).
Ecology and Pathogenesis
M. artica was first isolated from samples of sea-ice taken from the Canadian Basin in the Arctic Ocean. It grows optimally at a temperature of 25–27°C, a pH of pH 6.0–9.0, and a NaCl concentration of 0–12 %. It can also tolerate much lower temperatures down to 0°C and higher salinity in order to survive in the Arctic Ocean. (Zhang et al., 2008).
While not much is understood about M. artic's ecological role or the function of Marinomonas genus in general, we do know a few things. For one, M. artic is capable of degrading organic materials to be taken up by smaller organisms, however, it does not reduce nitrate nor does it produce hydrogen sulfide (Zhang et al. 2008).
Marinomonas primoryensis, which shares phylogeny with Marinomonas artica, has been proven to have IBPs (ice-binding proteins) that allow it to latch onto the sea ice and remain closer to the surface in aerobic and nutrient-rich conditions. IBP-producing bacteria are also known to reside in larger organisms and provide resistance to ice damage and freezing, though this is not the case for Marinomonas primoryensis or Marinomonas artica specifically. While research on the presence of IBPs has not been studied in M. artica, the presence of an adhesion protein that contains AFP (putative antifreeze protein) has been discovered in its genome. This AFP acts similarly to IBPs and provides insight as to how M. artica survives in its environment (Dolev et al., 2016; Liao et al,. 2021).
There has been no evidence to suggest pathogenic properties or symbiosis.
References
Chunming, D., Xiuhua, B., Qiliang, L., Yarong, X., Xin, C., and Zongze, S. "Draft Genome Sequence of Marinomonas sp. Strain D104, a polycyclic Aromatic Hydrocarbon-Degrading Bacterium from the Deep-Sea Sediment of the Artic Ocean." American Society for Microbiology Journals. 2014. https://doi.org/10.1128/genomeA.01211-13
Dolev, M.B., Bernheim, R., Guo, S., Davies, P.L., Braslavsky, I. "Putting life on ice: bacteria that bind to frozen water." The Royal Society. 2016. 13:12. https://doi.org/10.1098/rsif.2016.0210
"Genome assembly ASM1462346v1." Polar Institute of China. Sept. 16, 2020. https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_014623465.1/
Liao, L., Gao, S., Xu, Y., Su, S., Wen, J., Chen, B. "Complete genome sequence of Marinomonas arctica BSI20414, a giant antifreeze protein-producing bacterium isolated from Arctic sea ice." ScienceDirect. 2021. 57, 100829. https://doi.org/10.1016/j.margen.2020.100829
Lucas-Elío, P., Goodwin, L., Woyke, T., Pitluck, S., Nolan, M., Kyrpides, et al. "Complete genome sequence of Marinomonas posidonica type strain (IVIA-Po-181T). Sttandards in Genomic Science. 2012. 7:1, 31–43. doi: 10.4056/sigs.2976373
Yong, Y., Huirong, L., Yinxin, Z., and Bo, C. "Extracellular enzymes of cold-adapted bacteria from Artic sea ice, Canada Basin". Polar Biol. 2009. 32, 1539–1547. https://doi.org/10.1007/s00300-009-0654-x
Zhang DC, Li HR, Xin YH, Liu HC, Chen B, Chi ZM, Zhou PJ, Yu Y. Marinomonas arctica sp. nov., a psychrotolerant bacterium isolated from the Arctic. Int J Syst Evol Microbiol. 2008 Jul;58(Pt 7):1715-8. doi: 10.1099/ijs.0.65737-0. PMID: 18599722.
Author
Page authored by Holly Stogner, student of Prof. Bradley Tolar at UNC Wilmington.