Methanosphaerula palustris: Difference between revisions
| Line 11: | Line 11: | ||
|} | |} | ||
Methanosphaerula palustris | ''Methanosphaerula palustris'' | ||
==Description and Significance== | ==Description and Significance== | ||
Revision as of 13:32, 27 April 2012
Classification
Domain (Archaea); Phylum (Euryarchaeota); Class (Methanomicrobia); Order (Methanomicrobiales); most similar to the family Methanospirillaceae
Species
|
NCBI: Taxonomy |
Methanosphaerula palustris
Description and Significance
Methanosphaerula palustris is a recently described species details of which were generated from research conducted at Cornell University in 2009. This research reported detailed information regarding the species' phylogenetic placement, physiology/morphology, and habitat from which it was isolated. The type strain used in the study was given the designation E1-9cT. Researchers utilized culture, microscopy, and molecular based techniques to describe the species' characteristics [1].
Methanosphaerula palustris strain E1-9cT was isolated from a rich, minerotrophic fen in New York state in 2008. Methanosphaerula palustris strain E1-9cT was cultured using a modified PM1 medium which is used for culturing anaerobic microorganisms. While growing at a range of pH values, a pH of 5.5 was determined to be optimal for growth. Preferred temperatures for growth were optimal at 30 degrees Celsius, giving it appropriately a mesophilic designation. Methanosphaerula palustris is a methanogen (methane producing) which apparently utilizes hydrogen and carbon dioxide as electron donors and acceptors respectively, along with acetate as a carbon source giving it the classification of chemolithoheterotroph[1].
Interestingly, although Methanosphaerula palustris is a member of the order Methanomicrobiales, an order documented as having significant physiological variability among members within it. Phylogenetic analysis using 16 SrRNA and amino acid sequences indicates significant differences from other genera in this order, which has led to the proposition of Methanosphaerula as novel genus within Methanomicrobiales[1].
Genome Structure
The genome of Methanosphaerula palustris is about 2.92 million basepairs long with about 2866 predicted genes[8]. Also, the genome is composed of 2655 protein genes and 70 RNA genes. The chromosomes are circular and the GC content is 55.3%[7].
Cell Structure and Metabolism
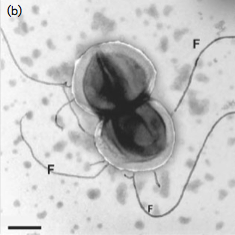
The cells of the regular cocci, Gram-positive Archaea are mostly associated in pairs, 0.5-0.8 mm in size. While multiple flagella are grown with 14 nm thick and 8–12 mm long. The flagella are readily lost under stress conditions such as high-speed centrifugation and oxic environments. The cell wall is about 30–70 nm thick and can be observed in thin sections under electron microscopy. As is shown in the study, cell walls can be resistant to SDS lysis, which indicates that it differs from single S-layer organisms; this is in spite of the fact that the S-layer is thought to be the only cell wall component in Archaea [1] [2].
It was shown that in Methanosphaerula palustris, methane is not produced from ethanol, methanol, 2-propanol, trimethylamine, 2-butanol, acetate, propionate or sodium butyrate. Only H2/CO2 and formate are used as substrates for methanogenesis under anaerobic conditions. Na2S is used as a sulfur source. Growth can be inhibited with formate concentrations above 50 mM or Na2S concentrations above 0.1 mM [1].
Ecology
Like other members of the order Methanomicrobiales, M. palustris inhabits peat forming wetlands, more specifically fens. These habitats occur sporadically in areas throughout the northern U.S and Canada. While conditions in these bog-like wetlands are typically acidic, fens are usually more alkaline, being fed through mineral-rich surface or groundwaters. These anaerobic, cool environments have fostered the buildup of large stores of organic carbon over thousands of years; peatlands contribute significantly to the global carbon pool. Additionally, these habitats are understood to contribute significantly to the global flux of methane (CH4) [3], due in part to the metabolic action of methanogenic microbes such as M. palustris. Methane is the most abundant organic compound found in the atmosphere, and a potent greenhouse gas. The EPA lists methane as having 21 C02 equivalents[4], giving it a relatively high global warming potential (GWP). Drastic increases in atmospheric methane over the past several hundred years have put it at its highest estimated atmospheric concentration in over 400,000 years although this trend is largely attributed to human activitiy [5]. Commercially, these bogs are also important sources of 'peat-moss'- a plant material that, after processing is slightly acidic, and capable of holding many times its weight in water making it an attractive component of soilless media used extensively in the horticultural industry.
References
[1]Cadillo-Quiroz, H et al. 2009. Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. International Journal of Systematic and Evolutionary Microbiology 59: 928–935.
[2]Wikipedia http://en.wikipedia.org/wiki/S-layer
[3}Harriss, R. et al. Methane flux from northern peatlands. Nature 315, 652 - 654 (20 June 1985); doi:10.1038/315652a0
[4]Environmental Protection Agency. <epa.gov/climatechange/glossary.html>
[5]National Research Council. "Summary." Climate Change Science: An Analysis of Some Key Questions. Washington, DC: The National Academies Press, 2001. 1. Print.
[6]http://en.wikipedia.org/wiki/fen
[7]http://www.genome.jp/kegg-bin/show_organism?org=mpl
[8]http://archaea.ucsc.edu/cgi-bin/hgGateway?db=methPalus1
Author
Page authored by Aaron Yoder, Shuya Zhang, and Jingyu Zhang, student of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->