Methylothermus thermalis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 87: | Line 87: | ||
<br> | <br> | ||
==References== | ==References== | ||
Revision as of 14:59, 21 April 2008
Classification
Bacteria; Proteobacteria; Gammaproteobacteria; Methylococcales; Methylococcaceae
Species
|
Methylothermus thermalis |
|
NCBI: Taxonomy |
Description and Significance
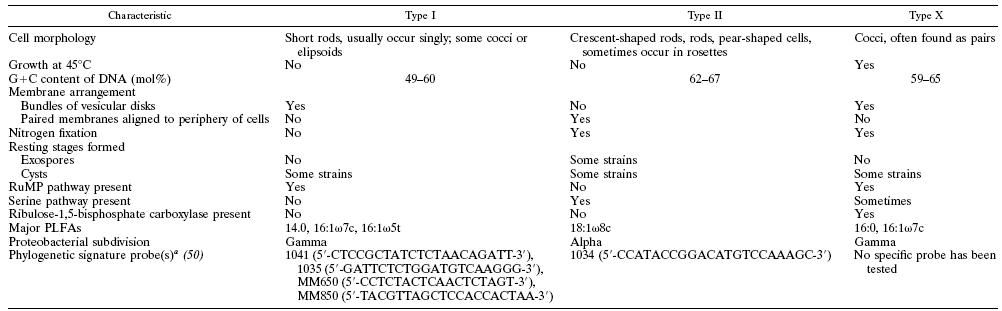
This organism was found in a Japanese hot spring (Hyogo, Japan). Methylothermus thermalis grew as small (1–2 mm in diameter), white, semi-transparent colonies with an entire edge and smooth surface were obsevered after 1-2 weeks. Older colonies (1 month) were light brown and more rigid. Unlike most discovered methanotrophs, this species can survive at high temperatures while metabolizing methane and methanol. It is classified as a type I thermophilic methanotroph, which uses the ribulose monophosphate (RuMP) pathway instead of the type II serine pathway. Methane monooxygenases present in aerobic methanotrophic bacteria exhibit a striking lack of substrate specificity, resulting in the fortuitous metabolism of a very large number of compounds including xenobiotic chemicals. Because of the ability of methanotrophs to catalyze a large number of biotransformations, they have attracted the interest of scientists involved in bioremediation and for the production of chemicals with commercial value.
Genome Structure
The nifH gene, which codes for nitrogenase reductase, was not detected with PCR analysis. Since nitrogenase reductase is component II of the nitrogenase enzyme complex, M. thermalis cannot fix its own atmospheric nitrogen. Also, the mmoX gene was not detected by PCR using the respective primer set that is universal for the known mmoX genes of methanotrophs. The genome of this microorganism has yet to be sequenced and as a consequence the size and characteristics are not well known.
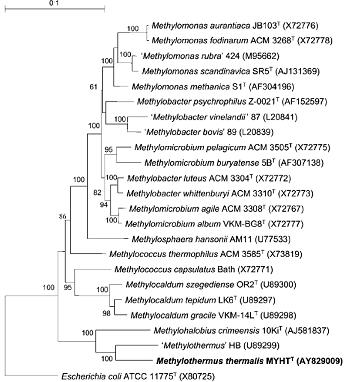
The G+C DNA content of 62.5 mol% for M. thermalis is close to that found for type II and type X methanotrophs. The 16S rRNA gene database search showed that this organism is closely related to the thermophilic methanotroph Methylothermus (91% sequence identity, based on the comparison of 1442 available bases) and the novel halophilic methanotroph Methylohalobius crimeensis (90 %). The other closest neighbors were the moderately thermophilic methanotrophs Methylococcus thermophilus and Methylocaldum szegediense, and the thermotolerant Methylococcus capsulatus Bath (about 85–86% 16S rRNA gene sequence identity). Phylogenetic analysis based on partial pmoA gene sequences confirmed the clustering of M. thermalis with Methylothermus strain HB (90% nucleotide sequence identity and 98% derived amino acid sequence identity) and Methylohalobius crimeensis (83 and 92 %, respectively). In contrast, the partial pmoA gene sequence of M. thermalis exhibited no more than 77% identity to the corresponding gene fragments of Methylocaldum szegediense, Methylocaldum gracile and Methylococcus capsulatus Bath. So far, information on true thermophilic methanotrophs is limited, with only a brief and non-formal taxonomic description of Methylothermus strain HB.
Analysis of the 16S rRNA and pmoA genes clearly showed that M. thermalis, together with Methylothermus strain HB and Methylohalobius crimeensis, represent a deepbranching group of extremophilic methanotrophs, which might indicate their common origin and consequent divergence. Therefore, M. thermalis appears to be more closely related to the type I (mesophilic, halophilic methanotroph Methylohalobius) than to the type X (thermophilic/tolerant Methylococcus and Methylocaldum) methanotrophs.
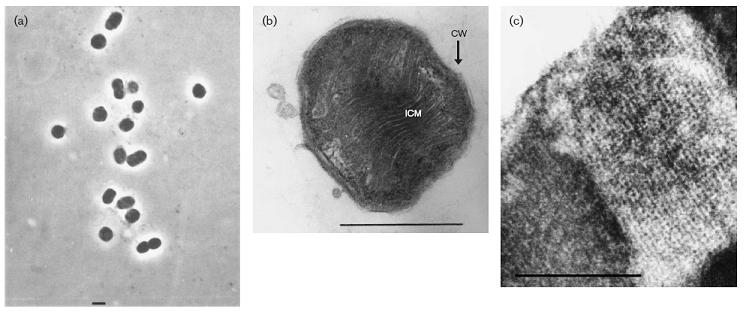
Cell Structure, Metabolism and Life Cycle
The cells grown in liquid culture were mostly represented by coccoids of 0.6–0.8 mm in diameter (Fig. 1a). In contrast, the majority of cells grown on solid medium appeared as short rods, slightly varying in size (0.6–0.8 x 1.0–1.2 mm). The cells were non-motile, and multiplied by normal cell division. The cells showed a typical Gram-negative structure of the cell wall, and the presence of type I intracytoplasmic membrane (ICM) arranged as stacks of vesicular disks (Fig. 1b) that completely filled the cytoplasmic space. Glycogen inclusions and poly-beta-hydroxybutyrate granules usually present in mesophilic methanotrophs were not observed. M. thermalis contained specific fatty acids, consisting mainly of C16 : 0 (37.24 %) and C18 : 1v9c (35.16 %). This organism's major polar lipids were phosphatidylethanolamine and phosphatidylglycerol. The major ubiquinone present was Q-8.
Methylothermus thermalis requires methane or methanol as a carbon and energy source to growth. This species can use nitrate, ammonia, urea, tryptophan,lysine, glutamate, formamide and Triss as nitrogen sources, but no growth was observed without one of these sources.
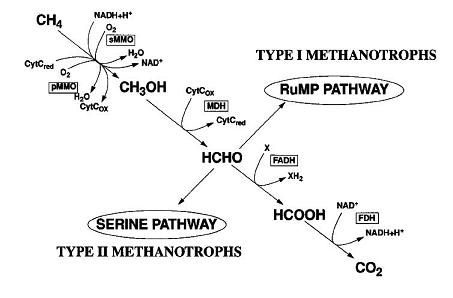
The oxidation of methane by aerobic methanotrophs is initiated by methane monooxygenases (MMOs), which are classical monooxygenases that utilize two reducing equivalents to split the O-O bonds of a dioxygen. One of the oxygen atoms is reduced to form H2O, and the other is incorporated into methane to form CH3OH. Two forms of MMOs have been found in methanotrophic bacteria. One form, a soluble MMO(sMMO) utilizes NADH + H+ as an electron donor. The sMMO does not contain heme cofactors or any other cofactors encountered previously in oxygenase chemistry. All known methanotrophs are capable of forming a particulate or membrane-bound MMO (pMMO). Extensive intracytoplasmic membranes and pMMO activity are present only when copper concentrations exceed 0.85 to 1 mmol/g (dry weight) of cells however.
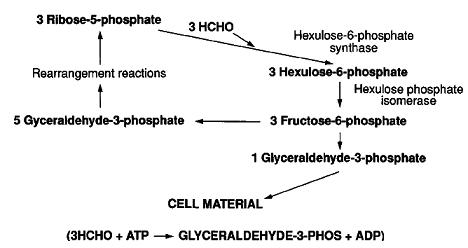
In the RuMP pathway, formaldehyde is combined with ribulose monophosphate via an aldol condensation to form hexulose-6-phosphate in a reaction catalyzed by hexulosephosphate synthase (HPS). Hexulose-6-phosphate is converted to fructose-6-phosphate by hexulosephosphate isomerase (HPI). These two enzymes are unique to the metabolism of one-carbon compounds by type I and type X methanotrophs and some other methylotrophs that do not utilize methane.
No activity of the key enzyme of the Calvin cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase, was found in this organism. The appropriate cbbL gene was not revealed by PCR. The complete tricarboxylic acid cycle does not operate in this organism because of the absence of a-ketoglutarate dehydrogenase. Like many other methanotrophs, the pyrophosphatedependent, but not the ATP-dependent, 6-phosphofructokinase was operative. Also, two possibilities for the cleavage of hexosephosphates exist, via fructose-1,6-bisphosphate aldolase and 2-keto-3-deoxy-6-phosphogluconate aldolase. The presence of glutamine synthetase and glutamate synthase activities indicated the involvement of the glutamate cycle in ammonia assimilation.
Ecology
The cells grew on methane or methanol at 37–67 6C, and optimally at 57–59 6C. They preferred media containing 0.5–1% NaCl and are capable of growth on 3% NaCl. M. thermalis is a neutrophile, growing at pH 6.5–7.5.
This organism converts methane and methanol into carbon dioxide aerobically; This species is a methanotroph and therefore it helps to sequester methane from the atmosphere. It converts this methane into carbon dioxide. Methane is a much more potent greenhouse gas, over 20 times more effective at trapping heat in the atmosphere, making this an attractive process to those worried about the increasing greenhouse effect. As an aerobe, this organism is able to take methane directly from the atmosphere and eat it up. Methanotrophs as a group are large players in the methane cycle on earth.
References
Author
Page authored by Anthony Heidt and James Goodrow Jr., student of Prof. Jay Lennon at Michigan State University.
