Mycoplasma haemocanis
Classification
Bacteria; Mycoplasmatota; Mollicutes; Mycoplasmatales; Mycoplasmataceae [5]
Species
|
NCBI: Taxonomy |
Mycoplasma haemocanis
Description and Significance
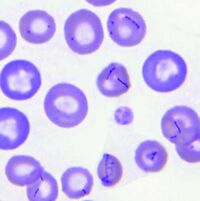
Mycoplasma haemocanis is a hemotrophic, gram-negative bacterial species apart of the Mycoplasma genus. The species and genus is characterized by its lack of a cell wall, lack of flagella, and its small size of less than 1 µm. Formerly known as Haemobartonella canis, this species was first discovered in Germany in 1928. It was later reclassified as a hemotrophic mycoplasma due to the similarity of its 16s ribosomal RNA (rRNA) gene and the 16s rRNA sequence of the Mycoplasma genus [4]. This species is commonly associated with causing hemolytic anemia in immunocompromised canines, specifically those who have undergone a splenectomy [1]. M. haemocanis attaches to the surface of erythrocytes (red blood cells) in hosts and can be visualized in a blood smear either as single coccoid cells or "bowed-looking" chains. This species is uncultivable in vitro, and instead depends on a host cell for survival [2,4]. The bacteria has a similar resemblance to the feline equivalent Mycoplasma haemofelis, which causes hemolytic anemia in cats. In a genetic comparison of the species' genomes, the two are actually concluded to be two different species infecting two distinct animals [2,3].
Genome Structure
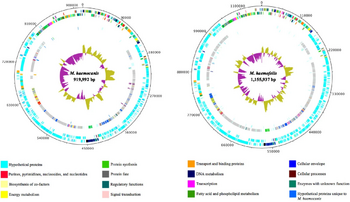
In 2012, one strain of M. haemocanis (Illinois) was isolated from the blood of an infected dog, then fully sequenced. The genome of the strain contains a singular circular chromosome comprising of 919,992 base pairs [2]. Compared to the average Mycoplasma genome size range of 580-2000 kb, the genome of M. haemocanis is consistent among other species in the same genus [4]. The genome also contains a low G+C (guanine and cytosine) content of 35%, which is characteristic for mycoplasmal species. Researchers discovered the presence of 1173 unique coding sequences (CDS), with around 75% coding for hypothetical proteins. Genome analyses of the CDS showed that 35% of the sequences are associated with the plasma membrane [2].
In comparing the genomes of M. haemocanis and M. haemofelis, analysis of the 16s rRNA sequence is not sufficient enough to distinguish whether the two species are different. Before 2012, the only genotypic evidence for the bacteria being separate species was the difference in RNase P gene sequences. Since the complete genome sequencing of the Illinois strain, the full sequence analysis including average nucleotide identity (ANI;85%) and tetranucleotides (0.95) has fallen below the cutoff value for being the same species [2].
Cell Structure, Metabolism and Life Cycle
Mycoplasma haemocanis features a relatively simple cell structure, lacking a cell wall. M. haemocanis most commonly appears morphologically as separate chains of cocci that are arranged linearly and adhere to the surface of red blood cells (cite Thompson). It can also appear as rod or ring-like structures. The absence of a cell wall causes the bacteria to stain gram negative.
It is a hemotrophic bacteria that relies on the host for the nutrients required for survival. The bacteria itself, like most hemoplasma, lacks the metabolic pathways that would be required for the production of energy as well as the synthesis of fundamental cellular components. This is likely a result of its adaptation to the blood habitat which is very nutrient rich [2]. M. haemocanis likely relies on metabolic pathways similar to the ones that M.haemofelis, its feline infecting counterpart, does as it features orthologs for all of the coding sequences identified in M. haemofelis. The bacteria is entirely dependent on erythrocytes and their growth factors but the exact methodology behind this is still widely unknown (CITE MILLAN). Mycoplasma haemocanis is able to thrive from exploiting the metabolism of red blood cells and garnering their nutrients; however, this comes at a cost to the erythrocytes themselves. The red blood cells experience a shortened lifespan and more intense anemia symptoms in the presence of M.haemocanis [2].
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
[1] Willi, B., Novacco, M., Meli, M., Wolf-Jäckel, G., Boretti, F., Wengi, N., Lutz, H., & Hofmann-Lehmann, R. (2010). Haemotropic mycoplasmas of cats and dogs: transmission, diagnosis, prevalence and importance in Europe. Schweizer Archiv fur Tierheilkunde, 152(5), 237–244. https://doi.org/10.1024/0036-7281/a000055
[2] do Nascimento, N.C., Santos, A.P., Guimaraes, A.M. et al. (2012). Mycoplasma haemocanis – the canine hemoplasma and its feline counterpart in the genomic era. Veterinary Research, 43(1), 66. https://doi.org/10.1186/1297-9716-43-66
[3] Hemotropic Mycoplasma infections in animals - circulatory system. Merck Veterinary Manual. (n.d.). https://www.merckvetmanual.com/circulatory-system/blood-parasites/hemotropic-mycoplasma-infections-in-animals
[4] Messick, J. B. (2004). Hemotrophic mycoplasmas (hemoplasmas): A review and new insights into pathogenic potential. Veterinary Clinical Pathology, 33(1), 2–13. https://doi.org/10.1111/j.1939-165x.2004.tb00342.x
[5] U.S. National Library of Medicine. (n.d.). Mycoplasma Haemocanis - NCBI - NLM. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/datasets/taxonomy/136241/
[6] Millán J, Di Cataldo S, Volokhov DV, Becker DJ. Worldwide occurrence of haemoplasmas in wildlife: Insights into the patterns of infection, transmission, pathology and zoonotic potential. Transbound Emerg Dis. 2021; 68: 3236–3256. https://doi.org/10.1111/tbed.13932
Author
Page authored by Erin Byers & Olivia Choros, students of Prof. Jay Lennon at IndianaUniversity.