Mycoplasma pneumoniae: Difference between revisions
No edit summary |
No edit summary |
||
| (18 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
[[Image:Mycoplasma pneumoniae.gif|frame|300px|right|Mycoplasma pneumoniae with permission to use granted by PROKARIOTAE Barros]] | [[Image:Mycoplasma pneumoniae.gif|frame|300px|right|Mycoplasma pneumoniae with permission to use granted by PROKARIOTAE Barros]] | ||
==Classification== | ==Classification== | ||
Bacteria; Firmicutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma pneumoniae | Bacteria; Firmicutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; "Mycoplasma pneumoniae" [[http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI]] | ||
==Description and significance== | |||
''Mycoplasma pneumoniae'' is a parasitic bacterium that invades the mucosal membranes of the upper and lower respiratory tract.(2) Mycoplasmas in general are bacteria that lack a cell wall, so they require residence in a host organism, such as a human or animal for survival.(2) These bacteria are spread through respiration and causes infections such as tracheobronchitis and primary atypical pneumonia. (2) The genome of ''M. pneumoniae'' is rather small only 600kb to 2300kb and encodes about 700 proteins. (1) Sequencing the genome of this bacteria gives an insight into the minimal amount of genetic material that is necessary of an organism to still be self sustaining and replicating. | |||
==Genome structure== | |||
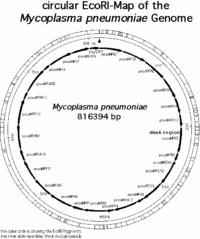
The genome of ''M. pneumoniae'' is rather small it contains only about 800kb and encodes 700 proteins. (1) The map of all the genes in the ''M. pnueomoiae'' genome had been identified by researchers in Germany in the laboratory of Richard Herrmann and are mapped out in the following pictures. | |||
[[Image:Ecomap.gif|thumb|200px|left|Mycoplasma pneumoniae chromosome mapped using EcoRI. Permission to use granted by Richard Herrmann: form Genome Project Website]] | |||
[[Image:Complete_Genome.gif|thumb|200px|right|Mycoplasma pneumoniae genome. Permission to use granted by Richard Herrmann: form Genome Project Website]] | |||
Based on the research conducted in the Genome Project by Richard Herrmann it was identified that the genome size of ''M. pneumoniae'' is 816394bp long, it has 677 open reading frames (ORF’s) corresponding to 716176 bp, the average MP-orf size is 39500 Da, and there are 39 (7998 bp) other coding regions for molecular machinery such as rRNA, tRNA, etc… (1) Overall the length of all the coding regions is 724174 bp, and that corresponds to 88.7 % of the entire genome. (1) The genome is contained in a double stranded circular chromosome, which has the highest G+C (40%) content of all the Mollicutes. (1,2,3) | |||
It is believed that ''M. pneumoniae'' developed from gram-positive bacteria and eventually through evolution lost much of its genetic material that would encode a cell wall and allow it to survive in the environment, and thus acquired a parasitic life style. (3) Using modern day molecular techniques and databases it is possible to identify genes that may be responsible for the pathogenesis of mycoplasmas and other pathogenic bacteria. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
The cell structure of | The cell structure of ''M. pneumoniae'' is filamentous or spherical and is like of all mycoplasmas, doesn’t contain a cell wall. (3) Because of this structural feature it can’t survive outside of a host due to osmotic instability in the environment. Therefore, ''M. pneumoniae'' is classified as a pathogen and parasite. (2,3) There are many challenges associated with treating infections and symptoms caused by this bacterium, since β-lactam antibiotics (ie. penicillin) can’t be used to kill the bacteria, because they target the lyses of the cell wall. (2) The membrane that ''M. pneumoniae'' has consists of sterols. (3) The outside membrane has an attachment organelle and many genes in the genome encode for production of these attachment organelles to ensure survival. (2,3) | ||
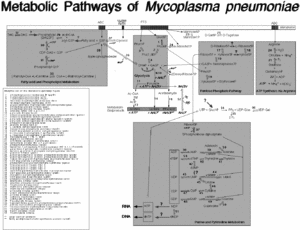
For M. pneumoniae to survive they need essential nutrients and compounds that include: amino acids, cholesterol, precursors to nucleic acid synthesis, and fatty acids, and can obtain all the essential compounds from the mucosal epithelial cells of the host. (1) Several metabolic pathways have been studied in the genome project of M. | For ''M. pneumoniae'' to survive they need essential nutrients and compounds that include: amino acids, cholesterol, precursors to nucleic acid synthesis, and fatty acids, and can obtain all the essential compounds from the mucosal epithelial cells of the host. (1) Several metabolic pathways have been studied in the genome project of ''M. pneumoniae'' by Richard Herrmann which include “ATP synthesis via Argenine,” “Sugar and central intermediae metabolism,” “Purine and Pyrimidine metabolism,” and “Fatty Acid metabolism.” The intricate and complex pathways of ''M. pneumoniae'' have been composed during the genome project for ''M. pneumoniae''. | ||
[[Image:Metabolic_Pathways.gif|thumb|300px|center|Mycoplasma pneumoniae Metabolic Pathways. Permission to use granted by Richard Herrmann: form Genome Project Website]] | |||
==Ecology== | ==Ecology== | ||
No Mycoplasma | No ''Mycoplasma pneumonae'' have been found to grow in the environment since they lack a cell wall and depend on the host for nutrient exchange and survival. (2) However, researchers have been able to devise methods and a medium rich enough to culture ''M. pneumoniae'' in the lab to be able to study it outside the host. (2) On agar, the bacteria grow in characteristic egg shape colonies. | ||
==Pathology== | ==Pathology== | ||
Mycoplasma pneumoniae is a parasitic bacterium that occupies the surface of the epithelial cells in the respiratory tact of its hosts and very often-causes pneumonia. (2,3) It attaches to the tracheal epithelial cells by sialoglycoproteins or sialoglycolipid receptors that are located on the surfaces of the cells using its attachment organelle, or adhesion proteins. (2,3) There have been many adhesion proteins discovered on the bacterial surfaces, among the most important are: P1 and P30. (3) Also it has been discovered by researchers that a vast extent of the bacterial genome contains sequences that encode for these adhesion proteins, suggesting that these proteins are very important for ensuring bacterial survival in the host. (3) Another very important feature of the bacteria that makes it a good pathogen and ensures its survival, is the lack of a cell wall around its periphery. Shmuel Razin, says that | ''Mycoplasma pneumoniae'' is a parasitic bacterium that occupies the surface of the epithelial cells in the respiratory tact of its hosts and very often-causes pneumonia. (2,3) It attaches to the tracheal epithelial cells by sialoglycoproteins or sialoglycolipid receptors that are located on the surfaces of the cells using its attachment organelle, or adhesion proteins. (2,3) There have been many adhesion proteins discovered on the bacterial surfaces, among the most important are: P1 and P30. (3) Also it has been discovered by researchers that a vast extent of the bacterial genome contains sequences that encode for these adhesion proteins, suggesting that these proteins are very important for ensuring bacterial survival in the host. (3) Another very important feature of the bacteria that makes it a good pathogen and ensures its survival, is the lack of a cell wall around its periphery. Shmuel Razin, says that | ||
“the lack of a cell wall in mycoplasmas may facilitate the direct contact of the mycoplasma membrane with that of its eukaryotic host, creating a condition which, in principle, could lead to fusion of the membranes, enabling the transfer or exchange of membrane components, and injection of the mycoplasmal cytoplasmic content, including hydrolytic enzymes, into the host cell cytoplasm. Thus, the potent nuclease of mycoplasmas combined with superoxide radicals may be responsible for clastogenic effects and chromosomal aberrations observed in | “the lack of a cell wall in mycoplasmas may facilitate the direct contact of the mycoplasma membrane with that of its eukaryotic host, creating a condition which, in principle, could lead to fusion of the membranes, enabling the transfer or exchange of membrane components, and injection of the mycoplasmal cytoplasmic content, including hydrolytic enzymes, into the host cell cytoplasm. Thus, the potent nuclease of mycoplasmas combined with superoxide radicals may be responsible for clastogenic effects and chromosomal aberrations observed in eukaryotic cells infected by mycoplasmas.” (Razin, p. 368) (3) | ||
The major damage that this adhesion causes is though to be due to the production of hydrogen peroxide (H2O2) and superoxide radicals (O2-) which are highly reactive species that cause host tissue damage.(3) Pneumonia is induced by the activation of macrophages and the production of immunoglobulins IgM, IgG, and IgA. (2) | The major damage that this adhesion causes is though to be due to the production of hydrogen peroxide (H2O2) and superoxide radicals (O2-) which are highly reactive species that cause host tissue damage.(3) Pneumonia is induced by the activation of macrophages and the production of immunoglobulins IgM, IgG, and IgA. (2) | ||
“Current extensive research of the modulation of the immune system by mycoplasmas appears to point out that the pathogenic manifestations of mycoplasma infections are in fact the outcome of the host immune reactions mediated by the production of various cytokines induced by the mycoplasmas, including the proinflamatory cytokines TNF-α, IL-1 and IL-6.” (Razin, p.368) (3) | “Current extensive research of the modulation of the immune system by mycoplasmas appears to point out that the pathogenic manifestations of mycoplasma infections are in fact the outcome of the host immune reactions mediated by the production of various cytokines induced by the mycoplasmas, including the proinflamatory cytokines TNF-α, IL-1 and IL-6.” (Razin, p.368) (3) | ||
The people most affected by pneumonia are young adults and children. The symptoms that one experienced after infection with M. pneumoniae are: cough, sore throat, low-grade fever, middle ear discomfort. (2) Many tests have been devised to diagnose whether the pneumonia is caused by M. pneumoniae or by something else. These tests include cold agglutinins, enzyme immunoassay, complete blood cell counts, erythrocyte sedimentation rates, and chest radiographs among others. (2) Mycoplasma pneumoniae bacteria are responsive to compounds such as tetracylines and macrolides. (2) There has not been any vaccine developed against mycoplasmas and the there is no immunological memory obtained after an infection, since recurrence of infection has been observed. (2) | The people most affected by pneumonia are young adults and children. The symptoms that one experienced after infection with ''M. pneumoniae'' are: cough, sore throat, low-grade fever, middle ear discomfort. (2) Many tests have been devised to diagnose whether the pneumonia is caused by ''M. pneumoniae'' or by something else. These tests include cold agglutinins, enzyme immunoassay, complete blood cell counts, erythrocyte sedimentation rates, and chest radiographs among others. (2) ''Mycoplasma pneumoniae'' bacteria are responsive to compounds such as tetracylines and macrolides. (2) There has not been any vaccine developed against mycoplasmas and the there is no immunological memory obtained after an infection, since recurrence of infection has been observed. (2) | ||
==Current Research== | ==Current Research== | ||
''Mycoplasma pneumnoiae'' is an organism that is of much interest to researchers because they “are the smallest free-living, self-replicating bacteria and possess unusually small genomes” (5). Researchers were studying the effects of infection with ''M. pneumoniae'' and looking at things like “mortality, body and lymph node weight, mycoplasma antibody titer, and lung mycoplasma burden and histopathology at intervals after infectionon” (5) They used mouse models that were wild type, were deficient in mast cells (cells of the immune system), and as a control a strain of mice infected with dead Mycoplasma cells. (5) The results of the study showed that those mice deficient in mast cells were more likely to die after infection since they lose more weight than wild type mice after infection.(5) When researchers compared the levels of IgA and IgM mycoplasma specific antibodies, they found that the mast cell deficient strain had an earlier and larger production of these immunoglobulins as did the wild type strain. (5) Also they observed that the strain of mice infected with the dead mycoplasma cells didn’t differ in any measured parameters from the wild type strain. The mast cell deficient strain also developed “larger bronchial lymph nodes and progressive pneumonia and airway occlusion.” (5) Based on the results obtained by researchers, they concluded that mast cells were very important in producing an effective innate immune response against infection as well as for recovery. (5) | |||
Researchers also studied the way the immune system responds to infection with ''M. pneumoniae''. In particular, how the immunoglobulins affect the mycoplasmal adhesion protein, which it needs to attach to the host cell, and what the “influence (is) on mycoplasma viability, metabolism, and cytadsorption.” (6) The study was performed using tracheal epithelial cells from a hamster and the antiserum and purified antibodies specific to target the P1 protein located on the mycoplasma membrane. (6) The anti-P1 antibody prevented the bacterium from attaching to the host hamster cells by about 80% and hemadsorption was inhibited by about 90%. (6) The immunoglobulins didn’t affect the metabolism or viability of the bacterium. (6) | |||
Through out much of the research conducted on ''M. pneumoniae'' it has been discovered that the P30 protein on the surface of the bacterium is very important for attachment to the host cell and ultimately for its survival as a parasite. (7) But researchers wanted to know what effect the P30 protein has on “gliding motility on the basis of satellite growth and cell gliding velocity and frequency.” (7) Researchers created a mutant which had a non-functioning P30 protein and another mutant with the P30 protein that was shorter in length. (7) To the researchers’ surprise “producing an altered P30 was unexpectedly not fully wild type with respect to gliding. Complementation of mutant II-3 with recombinant wild-type and mutant alleles confirmed the correlation between gliding defect and loss or alteration in P30.” (7) Researchers concluded that they P30 protein does have an affect on ''M. pneumoniae'' gliding motility, it is different from the role that P30 plays as an attachment organelle. (7) | |||
==References== | ==References== | ||
1) Herrmann, Richard “Understanding the Biology of a ‘minimal cell’,” The M.pneumoniae Genome Project, Unviersity of Heidelberg Germany. Website: | 1) Herrmann, Richard “Understanding the Biology of a ‘minimal cell’,” The M.pneumoniae Genome Project, Unviersity of Heidelberg Germany. | ||
Website: | |||
http://www.zmbh.uni-heidelberg.de/M_pneumoniae/genome/Results.html | http://www.zmbh.uni-heidelberg.de/M_pneumoniae/genome/Results.html | ||
http://www.zmbh.uni-heidelberg.de/M_pneumoniae/research.html | http://www.zmbh.uni-heidelberg.de/M_pneumoniae/research.html | ||
2) Razin, Shmuel. Ed. Samuel Baron. “Mycoplasmas” Medical Microbiology, 4th Edition, University of Texas Medial Branch at Galveston. Online Book: | 2) Razin, Shmuel. Ed. Samuel Baron. “Mycoplasmas” Medical Microbiology, 4th Edition, University of Texas Medial Branch at Galveston. | ||
Online Book: | |||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books&cmd=search&doptcmdl=TOCView&term=m.+pneumoniae+AND+mmed%5Bbook%5D | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books&cmd=search&doptcmdl=TOCView&term=m.+pneumoniae+AND+mmed%5Bbook%5D | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=books | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=books | ||
3) Razin, Shmuel. “Adherence of Pathogenic Mycoplasmas to Host Cells,” Bioscience Reports, Vol 19 No. 5 1999 pp.367-372. | 3) Razin, Shmuel. “Adherence of Pathogenic Mycoplasmas to Host Cells,” Bioscience Reports, Vol 19 No. 5 1999 pp.367-372. | ||
http://www.springerlink.com/content/m23t808600v75834/fulltext.pdf | http://www.springerlink.com/content/m23t808600v75834/fulltext.pdf | ||
http://www.springerlink.com/content/m23t808600v75834/ | http://www.springerlink.com/content/m23t808600v75834/ | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10763804&dopt=Abstract | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10763804&dopt=Abstract | ||
4) Howard, Katherine J. “Mycoplasma pneumoniae: the mystery bug.” | 4) Howard, Katherine J. “Mycoplasma pneumoniae: the mystery bug.” | ||
http://s99.middlebury.edu/BI330A/projects/Howard/Mpneumoniae.html | http://s99.middlebury.edu/BI330A/projects/Howard/Mpneumoniae.html | ||
5) Xu, Xian, et. al. “Mast Cells Protect Mice from Mycoplasma pneumonia.” American Journal of Respiratory and Critical Care Medicine. 2006. Vol 137. p 219-225. | |||
http://ajrccm.atsjournals.org/cgi/content/full/173/2/219 | |||
6) Krause, Duncan C. and Baseman, Joel B. “Inhibition of Mycoplasma pneumoniae Hemadsorption and Adherence to Respiratory Epithelium by Antibodies to a Membrane Protein.” Infection and Immunity. 1983. Vol 39. p.1180-1186. | |||
http://iai.asm.org/cgi/content/abstract/39/3/1180?ijkey=e9c47b7989e0a566fff45abfa582a1e83edc4766&keytype2=tf_ipsecsha | |||
http://iai.asm.org/cgi/reprint/39/3/1180?ijkey=e9c47b7989e0a566fff45abfa582a1e83edc4766 | |||
7) Hasselbring, Benjamin, et.al. “Mutant Analysis Reveals a Specific Requirement for Protein P30 in Mycoplasma pneumoniae Gliding Motility.” Journal of Bacteriology. 2005.Vol 187. No18. p.6281-6289. | |||
http://jb.asm.org/cgi/content/full/187/18/6281 | |||
Edited by Yelena Pasternak student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano at UCSD. | Edited by Yelena Pasternak student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano at UCSD. | ||
KMG | |||
Latest revision as of 18:42, 22 April 2011
A Microbial Biorealm page on the genus Mycoplasma pneumoniae
Classification
Bacteria; Firmicutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; "Mycoplasma pneumoniae" [NCBI]
Description and significance
Mycoplasma pneumoniae is a parasitic bacterium that invades the mucosal membranes of the upper and lower respiratory tract.(2) Mycoplasmas in general are bacteria that lack a cell wall, so they require residence in a host organism, such as a human or animal for survival.(2) These bacteria are spread through respiration and causes infections such as tracheobronchitis and primary atypical pneumonia. (2) The genome of M. pneumoniae is rather small only 600kb to 2300kb and encodes about 700 proteins. (1) Sequencing the genome of this bacteria gives an insight into the minimal amount of genetic material that is necessary of an organism to still be self sustaining and replicating.
Genome structure
The genome of M. pneumoniae is rather small it contains only about 800kb and encodes 700 proteins. (1) The map of all the genes in the M. pnueomoiae genome had been identified by researchers in Germany in the laboratory of Richard Herrmann and are mapped out in the following pictures.
Based on the research conducted in the Genome Project by Richard Herrmann it was identified that the genome size of M. pneumoniae is 816394bp long, it has 677 open reading frames (ORF’s) corresponding to 716176 bp, the average MP-orf size is 39500 Da, and there are 39 (7998 bp) other coding regions for molecular machinery such as rRNA, tRNA, etc… (1) Overall the length of all the coding regions is 724174 bp, and that corresponds to 88.7 % of the entire genome. (1) The genome is contained in a double stranded circular chromosome, which has the highest G+C (40%) content of all the Mollicutes. (1,2,3) It is believed that M. pneumoniae developed from gram-positive bacteria and eventually through evolution lost much of its genetic material that would encode a cell wall and allow it to survive in the environment, and thus acquired a parasitic life style. (3) Using modern day molecular techniques and databases it is possible to identify genes that may be responsible for the pathogenesis of mycoplasmas and other pathogenic bacteria.
Cell structure and metabolism
The cell structure of M. pneumoniae is filamentous or spherical and is like of all mycoplasmas, doesn’t contain a cell wall. (3) Because of this structural feature it can’t survive outside of a host due to osmotic instability in the environment. Therefore, M. pneumoniae is classified as a pathogen and parasite. (2,3) There are many challenges associated with treating infections and symptoms caused by this bacterium, since β-lactam antibiotics (ie. penicillin) can’t be used to kill the bacteria, because they target the lyses of the cell wall. (2) The membrane that M. pneumoniae has consists of sterols. (3) The outside membrane has an attachment organelle and many genes in the genome encode for production of these attachment organelles to ensure survival. (2,3) For M. pneumoniae to survive they need essential nutrients and compounds that include: amino acids, cholesterol, precursors to nucleic acid synthesis, and fatty acids, and can obtain all the essential compounds from the mucosal epithelial cells of the host. (1) Several metabolic pathways have been studied in the genome project of M. pneumoniae by Richard Herrmann which include “ATP synthesis via Argenine,” “Sugar and central intermediae metabolism,” “Purine and Pyrimidine metabolism,” and “Fatty Acid metabolism.” The intricate and complex pathways of M. pneumoniae have been composed during the genome project for M. pneumoniae.
Ecology
No Mycoplasma pneumonae have been found to grow in the environment since they lack a cell wall and depend on the host for nutrient exchange and survival. (2) However, researchers have been able to devise methods and a medium rich enough to culture M. pneumoniae in the lab to be able to study it outside the host. (2) On agar, the bacteria grow in characteristic egg shape colonies.
Pathology
Mycoplasma pneumoniae is a parasitic bacterium that occupies the surface of the epithelial cells in the respiratory tact of its hosts and very often-causes pneumonia. (2,3) It attaches to the tracheal epithelial cells by sialoglycoproteins or sialoglycolipid receptors that are located on the surfaces of the cells using its attachment organelle, or adhesion proteins. (2,3) There have been many adhesion proteins discovered on the bacterial surfaces, among the most important are: P1 and P30. (3) Also it has been discovered by researchers that a vast extent of the bacterial genome contains sequences that encode for these adhesion proteins, suggesting that these proteins are very important for ensuring bacterial survival in the host. (3) Another very important feature of the bacteria that makes it a good pathogen and ensures its survival, is the lack of a cell wall around its periphery. Shmuel Razin, says that “the lack of a cell wall in mycoplasmas may facilitate the direct contact of the mycoplasma membrane with that of its eukaryotic host, creating a condition which, in principle, could lead to fusion of the membranes, enabling the transfer or exchange of membrane components, and injection of the mycoplasmal cytoplasmic content, including hydrolytic enzymes, into the host cell cytoplasm. Thus, the potent nuclease of mycoplasmas combined with superoxide radicals may be responsible for clastogenic effects and chromosomal aberrations observed in eukaryotic cells infected by mycoplasmas.” (Razin, p. 368) (3) The major damage that this adhesion causes is though to be due to the production of hydrogen peroxide (H2O2) and superoxide radicals (O2-) which are highly reactive species that cause host tissue damage.(3) Pneumonia is induced by the activation of macrophages and the production of immunoglobulins IgM, IgG, and IgA. (2)
“Current extensive research of the modulation of the immune system by mycoplasmas appears to point out that the pathogenic manifestations of mycoplasma infections are in fact the outcome of the host immune reactions mediated by the production of various cytokines induced by the mycoplasmas, including the proinflamatory cytokines TNF-α, IL-1 and IL-6.” (Razin, p.368) (3)
The people most affected by pneumonia are young adults and children. The symptoms that one experienced after infection with M. pneumoniae are: cough, sore throat, low-grade fever, middle ear discomfort. (2) Many tests have been devised to diagnose whether the pneumonia is caused by M. pneumoniae or by something else. These tests include cold agglutinins, enzyme immunoassay, complete blood cell counts, erythrocyte sedimentation rates, and chest radiographs among others. (2) Mycoplasma pneumoniae bacteria are responsive to compounds such as tetracylines and macrolides. (2) There has not been any vaccine developed against mycoplasmas and the there is no immunological memory obtained after an infection, since recurrence of infection has been observed. (2)
Current Research
Mycoplasma pneumnoiae is an organism that is of much interest to researchers because they “are the smallest free-living, self-replicating bacteria and possess unusually small genomes” (5). Researchers were studying the effects of infection with M. pneumoniae and looking at things like “mortality, body and lymph node weight, mycoplasma antibody titer, and lung mycoplasma burden and histopathology at intervals after infectionon” (5) They used mouse models that were wild type, were deficient in mast cells (cells of the immune system), and as a control a strain of mice infected with dead Mycoplasma cells. (5) The results of the study showed that those mice deficient in mast cells were more likely to die after infection since they lose more weight than wild type mice after infection.(5) When researchers compared the levels of IgA and IgM mycoplasma specific antibodies, they found that the mast cell deficient strain had an earlier and larger production of these immunoglobulins as did the wild type strain. (5) Also they observed that the strain of mice infected with the dead mycoplasma cells didn’t differ in any measured parameters from the wild type strain. The mast cell deficient strain also developed “larger bronchial lymph nodes and progressive pneumonia and airway occlusion.” (5) Based on the results obtained by researchers, they concluded that mast cells were very important in producing an effective innate immune response against infection as well as for recovery. (5)
Researchers also studied the way the immune system responds to infection with M. pneumoniae. In particular, how the immunoglobulins affect the mycoplasmal adhesion protein, which it needs to attach to the host cell, and what the “influence (is) on mycoplasma viability, metabolism, and cytadsorption.” (6) The study was performed using tracheal epithelial cells from a hamster and the antiserum and purified antibodies specific to target the P1 protein located on the mycoplasma membrane. (6) The anti-P1 antibody prevented the bacterium from attaching to the host hamster cells by about 80% and hemadsorption was inhibited by about 90%. (6) The immunoglobulins didn’t affect the metabolism or viability of the bacterium. (6)
Through out much of the research conducted on M. pneumoniae it has been discovered that the P30 protein on the surface of the bacterium is very important for attachment to the host cell and ultimately for its survival as a parasite. (7) But researchers wanted to know what effect the P30 protein has on “gliding motility on the basis of satellite growth and cell gliding velocity and frequency.” (7) Researchers created a mutant which had a non-functioning P30 protein and another mutant with the P30 protein that was shorter in length. (7) To the researchers’ surprise “producing an altered P30 was unexpectedly not fully wild type with respect to gliding. Complementation of mutant II-3 with recombinant wild-type and mutant alleles confirmed the correlation between gliding defect and loss or alteration in P30.” (7) Researchers concluded that they P30 protein does have an affect on M. pneumoniae gliding motility, it is different from the role that P30 plays as an attachment organelle. (7)
References
1) Herrmann, Richard “Understanding the Biology of a ‘minimal cell’,” The M.pneumoniae Genome Project, Unviersity of Heidelberg Germany.
Website:
http://www.zmbh.uni-heidelberg.de/M_pneumoniae/genome/Results.html
http://www.zmbh.uni-heidelberg.de/M_pneumoniae/research.html
2) Razin, Shmuel. Ed. Samuel Baron. “Mycoplasmas” Medical Microbiology, 4th Edition, University of Texas Medial Branch at Galveston.
Online Book:
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=books
3) Razin, Shmuel. “Adherence of Pathogenic Mycoplasmas to Host Cells,” Bioscience Reports, Vol 19 No. 5 1999 pp.367-372.
http://www.springerlink.com/content/m23t808600v75834/fulltext.pdf
http://www.springerlink.com/content/m23t808600v75834/
4) Howard, Katherine J. “Mycoplasma pneumoniae: the mystery bug.”
http://s99.middlebury.edu/BI330A/projects/Howard/Mpneumoniae.html
5) Xu, Xian, et. al. “Mast Cells Protect Mice from Mycoplasma pneumonia.” American Journal of Respiratory and Critical Care Medicine. 2006. Vol 137. p 219-225.
http://ajrccm.atsjournals.org/cgi/content/full/173/2/219
6) Krause, Duncan C. and Baseman, Joel B. “Inhibition of Mycoplasma pneumoniae Hemadsorption and Adherence to Respiratory Epithelium by Antibodies to a Membrane Protein.” Infection and Immunity. 1983. Vol 39. p.1180-1186.
http://iai.asm.org/cgi/reprint/39/3/1180?ijkey=e9c47b7989e0a566fff45abfa582a1e83edc4766
7) Hasselbring, Benjamin, et.al. “Mutant Analysis Reveals a Specific Requirement for Protein P30 in Mycoplasma pneumoniae Gliding Motility.” Journal of Bacteriology. 2005.Vol 187. No18. p.6281-6289.
http://jb.asm.org/cgi/content/full/187/18/6281
Edited by Yelena Pasternak student of Rachel Larsen and Kit Pogliano at UCSD.
KMG