Oil-Eating Bacteria: A Tool for Bioremediation
Introduction
The use of petroleum, or crude oil, has been increasing since the industrial revolution [1]. The two common uses of petroleum are fuel and the synthesis of organic compounds [2]. Petroleum fuel is used as an energy source to provide many common-day luxuries, at least in the first world, such as heating and lighting for buildings and homes and for transportation [2]. Starting in 2000, petroleum was used to synthesize 99% of human-made organic compounds such as gasoline and kerosine [2]. Clearly, we rely heavily on petroleum and it is a ubiquitous chemical in our daily lives; however, there are a plethora of environmental consequences to such high petroleum consumption levels.

Petroleum pollution is one of the most common pollutants in the world and is harmful to almost every species on Earth [4]. Each year, three million tons of petroleum enters the ocean through activities such as oil-pumping, transportation, and more than half coming from prohibited activities and accidents [3]. Numerous studies have documented the negative consequences of petroleum pollution on organismal life. For example, petroleum has been found to interfere with the photosynthesis pathway of phytoplankton, which are the primary producers (base of the food chain) in marine environments [5]. Therefore, decreases in phytoplankton abundance caused by petroleum pollution could have bottom-up effects on the whole marine ecosystem [5]. Coral reefs are also negatively affected by petroleum, and a study by Turner and Renegar (2017) found that petroleum exposure can cause decreases in the abundance and diversity of coral by reducing growth rates, reproduction rates, and increasing tissue loss and death. Not only are marine environments affected by petroleum pollution, but terrestrial environments can also be affected through soil and groundwater contamination [4]. Clean groundwater trapped in aquifers is a precious resource for humans, making the contamination of groundwater a serious issue [4].
Given the multiple consequences of petroleum pollution today, it appears that we desperately need an environmentally-friendly technique to remove petroleum from contaminated ecosystems. One such technique involves bioremediation, which is the use of microbes to convert toxic pollutants, such as anthropogenic petroleum, into non-toxic compounds [6]. Recent scientific work has found that some bacteria (Oleispira, Oleiphilus, Thalassolituus, Alcanivorax and Cycloclasticus) can feed on petroleum due to their hydrocarbon‐degrading abilities [3]. Evolution has played a role in the formation of species capable of degrading petroleum. Petroleum oil is naturally found in environments from seeps in the seafloor and from fatty acids and lipids produced by plants and animals [7]. To colonize these petroleum niches, some bacteria have evolved to produce enzymes capable of breaking down petroleum and using it as energy source for their metabolic pathways [7]. Therefore, there is hope in the scientific community that we can utilize oil-degrading bacteria and enhance their abilities through genetic engineering to create a bioremediation tool to remove anthropogenic petroleum from polluted environments.
Alcanivorax borkumensis
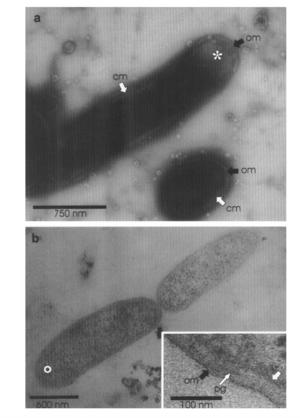
While multiple bacteria have the capacity to break down petroleum, Alcanivorax borkumensis appears to be the dominant bacteria among oil-degrading bacteria, making up 80% to 90% of the microbes in oil-polluted water [9]. A. borkumensis was first discovered in 1998 by a team of microbiologists led by Michail Yakimov [8]. The researchers isolated A. borkumensis from sea water and sediment samples in the North Sea, and through experimental analysis they found that the bacteria is a rod-shaped, gram-negative, non-spore-forming, non-motile, aerobic bacteria in the γ-proteobacterium phylum [8]. Since it was first discovered in 1998, A. borkumensis has been observed across the world, from the Pacific Ocean to the Arctic Sea [9]. The figure on the left is from electron microscopy imaging and shows A. borkumensis and its outer membrane [8], a requisite for gram-negative bacteria.
A. borkumensis grows slowly and can grow almost entirely on alkanes [9], which is one of the major compounds found in petroleum. In unpolluted water, A. borkumensis abundance is generally low; however, these bacteria become abundant in oil-polluted waters. For optimal growth, the bacteria requires a temperature between 25 and 30°C, but they can grow between 4 and 35°C [8]. Additionally, the necessary salinity concentration for A. borkumensis can be as low as 1% and as high as 12.5%, with the optimal salinity concentration ranging from 3 to 10% NaCl [8]. Yakimov and researchers found that A. borkumensis are 0.6-0.8 μm wide and 16-25 μm long if growing on pyruvate-supplemented medium, but are much shorter (1-1.5 μm) if n-alkanes are the sole carbon source. Additionally, the cell wall of A. borkumensis is generally 31 to 36 nm [8].
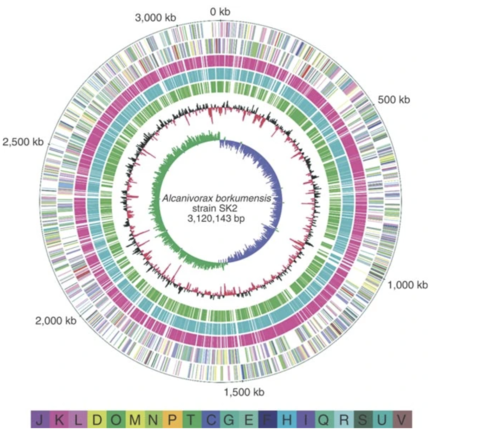
The key to A. borkumensis superior ability to degrade petroleum can be found in its genome, which contains a circular genome of 3,120,143 base pairs (bp) and a mean G+C content of 54.7% [9]. Within the genome of A. borkumensis are a plethora of different systems that help the bacteria attach itself to petroleum through pili and biosurfactants and degrade petroleum through enzyme formation [9]. For instance, A. borkumensis has 16 pili genes that help the bacteria attach themselves to oil in water. In addition, many secretion systems, such as the Type II secretion system, Sec translocon, twin-arginin translocation (Tat), and five genes from the HlyD family, are used by A. borkumensis to help them create biofilms, which are a cluster of microorganisms living on a surface [9]. After bacteria have attached themselves to the surface of petroleum, they need a way to degrade petroleum hydrocarbons. Within the genome of A. borkumensis is the alkSB1GHJ operon, which is known to contain genes for the alkane degradation pathway [9]. One important gene in this operon is the AlkB1 gene which makes an alkane hydroxylase that oxidizes medium-chain alkanes with 5 to 12 carbons (Schneiker et al., 2006). AlkB2 is another gene with the same function as AlkB1 but it hydrolyzes medium-chain alkanes with 8 to 16 carbons and it is encoded by a different alkane hydroxylase system (ABO_0122) [9]. By searching through databases, Schneiker and colleagues found that relatively few marine species have AlkB genes and even fewer have two types of this gene. In addition to the genes discussed above, A. borkumensis also has three cytochromes (ABO_2384 for P450, ABO_2288 P450, and ABO_0201 P450) that contain genes involved in alkane degradation [9]. Surprisingly, even more enzyme-encoding genes for alkane degradation, such as alkK, alkL, alkN, rubA, and rubB, were also found in the A. borkumensis genome [9]. Due to the wide range of alkane degradation pathways, A. borkumensis is able to efficiently break down petroleum compared to other species. The figure below shows the genes involved in alkane degradation (a) as well as a phylogenetic tree of bacteria with similar P450 cytochromes (b) and the possible alkane degradation pathway (c).

Section 3
Include some current research, with at least one figure showing data.
Biodegradation Mechanism
Include some current research, with at least one figure showing data.
By
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki.
The insertion code consists of:
Double brackets: [[
Filename: Biodegradation_of_Pollutants.png
Thumbnail status: |thumb|
Pixel size: |500px|
Placement on page: |right|
Legend/credit: Every image requires a link to the source.
Closed double brackets: ]]
Possible Use in Biotechnology
Conclusion
References
- ↑ Villela, H.D.M, Peixoto, R.S., Soriano, A.U., Carmo, F.L. (2019). Microbial bioremediation of oil contaminated seawater: A survey of patent deposits and the characterization of the top genera applied. Science of The Total Environment. 666: 743-758.
- ↑ 2.0 2.1 2.2 Eneh, O. C. (2011). A Review on Petroleum: Source, Uses, Processing, Products and the Environment. Journal of Applied Sciences. 11(12): 2084-2091.
- ↑ 3.0 3.1 3.2 Brooijmans, R. J., Pastink, M. I., & Siezen, R. J. (2009). Hydrocarbon-degrading bacteria: the oil-spill clean-up crew. Microbial biotechnology, 2(6), 587–594
- ↑ 4.0 4.1 4.2 Logeshwaran, P., Megharaj, M., Chadalavada, S., Bowman, M., Naidu, R.. (2018). Petroleum hydrocarbons (PH) in groundwater aquifers: An overview of environmental fate, toxicity, microbial degradation and risk-based remediation approaches. Environmental Technology & Innovation. 10: 175-193.
- ↑ 5.0 5.1 Jiang, Z., Huang, Y., Xu, X., Liao, Y., Shou, L., Liu, J., Chen, Q., Zeng, J. (2010).Advance in the toxic effects of petroleum water accommodated fraction on marine Plankton. Acta Ecologica Sinica. 30(1): 8-15.
- ↑ Ezezika, O.C. and Singer, P.A. (2010). Genetically engineered oil-eating microbes for bioremediation: Prospects and regulatory challenges. Technology in Society. 32(4): 331-335.
- ↑ 7.0 7.1 Yakimov, M.M. Timmis, K.N., Golyshin, P.N. (2007). Obligate oil-degrading marine bacteria. Current Opinion in Biotechnology. 18(3): 257-266.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Yakimov, Michail M.; et al. (1998). "Alcanivorax Borkumensis gen. nov., sp. nov., A New, Hydrocarbon-degrading And Surfactant-producing Marine Bacterium". International Journal of Systematic Bacteriology. 48 (2): 339–348
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 Schneiker, S., dos Santos, V., Bartels, D. et al. (2006) Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol 24, 997–1004.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2021, Kenyon College.
