Panteoa agglomerans: Difference between revisions
No edit summary |
No edit summary |
||
| Line 21: | Line 21: | ||
P. agglomerans demonstrates a wide array of medical applications as well, producing multiple compounds that are utilized to produce powerful biocontrol agents for humans and plants that function as antibiotics, antifungals, and antivirals. P. agglomerans also produces a unique lipopolysaccharide that acts as a macrophage activator and immunopotentiator which has been utilized in research to suppress cancerous tumors. | P. agglomerans demonstrates a wide array of medical applications as well, producing multiple compounds that are utilized to produce powerful biocontrol agents for humans and plants that function as antibiotics, antifungals, and antivirals. P. agglomerans also produces a unique lipopolysaccharide that acts as a macrophage activator and immunopotentiator which has been utilized in research to suppress cancerous tumors. | ||
[[File:P agglomerans uses.png|thumb| | [[File:P agglomerans uses.png|thumb|00px|center|Image credit: Walterson, A. M., & Stavrinides, J.,2015]] | ||
==Genome Structure== | ==Genome Structure== | ||
P. agglomerans has a single circular genome 4.8Mb long with a median protein count of 4,435 and a GC content of 55%. The chromosome contains 3-5 large plasmids ranging from 280-789 kb as well as 3,615 coding sequences, 77 tRNA genes and 22 rRNA operons. The genome has been fully sequenced to 76-fold coverage by a whole-genome shotgun strategy. One shotgun and 0.5 paired-end reads were performed using a Roche 454 GS (FLX titanium) pyrosequencer. All of the reads were assembled using Newbler Assembler 2.5 (454 Life Science), which generated 18 large contigs (>500 bp). | P. agglomerans has a single circular genome 4.8Mb long with a median protein count of 4,435 and a GC content of 55%. The chromosome contains 3-5 large plasmids ranging from 280-789 kb as well as 3,615 coding sequences, 77 tRNA genes and 22 rRNA operons. The genome has been fully sequenced to 76-fold coverage by a whole-genome shotgun strategy. One shotgun and 0.5 paired-end reads were performed using a Roche 454 GS (FLX titanium) pyrosequencer. All of the reads were assembled using Newbler Assembler 2.5 (454 Life Science), which generated 18 large contigs (>500 bp). | ||
Revision as of 23:34, 11 December 2023
Classification
Bacteria, Pseudomonadota, Gammaproteobacteria, Enterobacterales, Erwiniaceae, Pantoea
Species
|
NCBI: [1] |
Pantoea agglomerans
Description and Significance
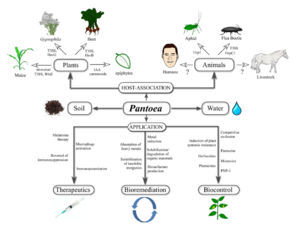
Pantoea agglomerans was first described in 1888 and has been identified under many names including: Enterobacter agglomerans, Erwinia herbicola, Bacillus agglomerans, Bacterium herbicola, Pseudomonas herbicola, Agrobacterium gypsophilae, Bacterium typhi flavum, Erwinia lathyr, Corynebacterium beticola, Flavobacterium rhenanum, Pseudomonas trifolii, and Xanthomonas trifolii. The etymology of its name is ‘Pantoea’, derived from the Greek word ‘pantoios’, which means ‘of all sorts and sources’, reflecting its ubiquitous occurrence; and ‘agglomerans’ meaning ‘forming into a ball’, due to the tendency for P. agglomerans to aggregate into clusters.
Pantoea agglomerans is a non-spore forming, Gram-negative, rod-shaped, facultative anaerobe bacillus that is motile via peritrichous flagella. It forms yellow pigmented colonies with 5mm concave centers. P. agglomerans grows quickly on agar at 30°C at a rate comparable to E. coli. P. agglomerans is found ubiquitously on plants as both an epiphyte and an endophyte. It forms host associations with a diverse variety of hosts including plants, insects, and humans. P. agglomerans is found abundantly in plants, animals, arthropods, feces, soil, water, dust, air, and in humans. P. agglomerans is as ubiquitous in function and application as it is in occurrence, it produces a wide variety of molecules depending on where it is grown with both positive and negative attributes and wide applications for bioremediation, biocontrol, and therapeutics. It has multiple bioremediation applications as a natural fertilizer and pesticide, replacing harmful chemical alternatives. Other bioremediation applications include natural feed additives for poultry, breaking down herbicides into nontoxic byproducts, and the ability to break down petroleum hydrocarbons. P. agglomerans demonstrates a wide array of medical applications as well, producing multiple compounds that are utilized to produce powerful biocontrol agents for humans and plants that function as antibiotics, antifungals, and antivirals. P. agglomerans also produces a unique lipopolysaccharide that acts as a macrophage activator and immunopotentiator which has been utilized in research to suppress cancerous tumors.
Genome Structure
P. agglomerans has a single circular genome 4.8Mb long with a median protein count of 4,435 and a GC content of 55%. The chromosome contains 3-5 large plasmids ranging from 280-789 kb as well as 3,615 coding sequences, 77 tRNA genes and 22 rRNA operons. The genome has been fully sequenced to 76-fold coverage by a whole-genome shotgun strategy. One shotgun and 0.5 paired-end reads were performed using a Roche 454 GS (FLX titanium) pyrosequencer. All of the reads were assembled using Newbler Assembler 2.5 (454 Life Science), which generated 18 large contigs (>500 bp). P. agglomerans has been identified as a cause of human infection by utilizing PCR techniques to identify the repA gene, which is linked to plant pathogenicity. P. agglomerans is symbiotic with locust species, producing hpc and vgrG, which code for effector proteins that make vital hormones. It also contains quorum sensing genes such as acyl-homoserine lactones.
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
Author
Page authored by Samuel Rogers, student of Prof. Bradley Tolar at UNC Wilmington. [Category:Pages edited by students of Bradley Tolar at UNC Wilmington]]