Pseudomonas corrugata: Difference between revisions
Cmcole1736 (talk | contribs) No edit summary |
Cmcole1736 (talk | contribs) No edit summary |
||
| Line 18: | Line 18: | ||
=16S Ribosomal RNA Gene Information= | =16S Ribosomal RNA Gene Information= | ||
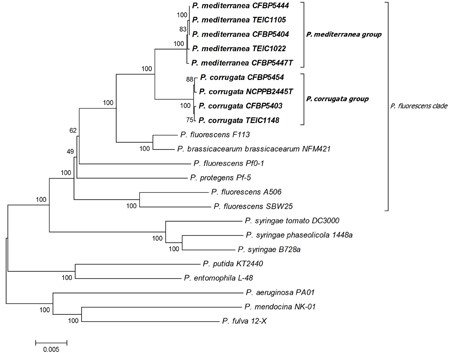
The closest relatives are: ''Pseudomonas brassicacearum'', with 97% similarity (Licciardello, et al, 2016). | |||
[[image:A1F1|thumbnail|400px|Figure 2: 16s rRNA sequence Image credit: Slabbink, et al]] | [[image:A1F1|thumbnail|400px|Figure 2: 16s rRNA sequence Image credit: Slabbink, et al]] | ||
| Line 27: | Line 27: | ||
Genome size 6,193,893 | Genome size is approximately 6,193,893, the DNA Coding Region has 4,577,116 base pairs, DNA G+C content 60.51%, Total Genes 6,211, Protein Coding Genes 6,165, Genes with Predicted Function 4,074. (Licciardello, et al, 2014) | ||
DNA Coding Region | |||
DNA G+C content 60.51% | |||
Total Genes 6,211 | |||
Protein Coding Genes 6,165 | |||
Genes with Predicted Function | |||
(Licciardello, et al, 2014) | |||
=Cell structure and metabolism= | =Cell structure and metabolism= | ||
Revision as of 21:19, 2 December 2016
Classification
Kingdom: Bacteria Phylum: Proteobacteria Class: Gammaproteobacteria Order: Pseudomonadales Family: Pseudomonadaceae Genus: 'Pseudomonas'
Species
Pseudomonas corrugata consists of at least 87 different strains (Catara, Sutra, Morineau, Achouak, Christen and Gardan, 2002) and there are at least 25 more species in the P. fluorescens group. In total there are at least 132 species total within the genus. There are also a total of 7 groups within the genus. The Pcor species epithet “corrugata” refers to the typical morphology of bacterial colonies on NDA; slightly raised, with a wrinkled-rough surface and undulate margins that produce a yellow diffusible pigment (Scarlett et al., 1978). This bacterium is one of the non-fluorescent Pseudomonas species.
Description and significance
P. corrugata was discovered in 1978 by Roberts and Scarlett (Scarlett, et al, 1978). This microbe is significant because uncontrolled outbreaks of it in agricultural communities can severely damage tomato crops for a season, having a huge economic impact. This bacteria attacks the pith of the tomato plant causing necrosis. Necrosis is rotting of the cell structure. P. corrugata produces wrinkled and, rarely, smooth colonies on yeast peptone glucose agar or nutrient dextrose agar; yellow to brown diffusible pigments are frequently produced. Disease symptoms include: typical symptom on tomato is necrosis and/or hollowing of the pith of the stem, the syndrome determines loss of turgidity of the plant, hydropic/necrotic areas, and long conspicuous adventitious roots on the stem. In vitro assessed against plant pathogenic fungi and bacteria, as well as the phytotoxin indicator microorganism Rhodotorula pilimanae and Bacillus megaterium; in vivo used against pre- and post-harvest plant pathogens (Catara, 2007).
16S Ribosomal RNA Gene Information
The closest relatives are: Pseudomonas brassicacearum, with 97% similarity (Licciardello, et al, 2016).
Genome Structure
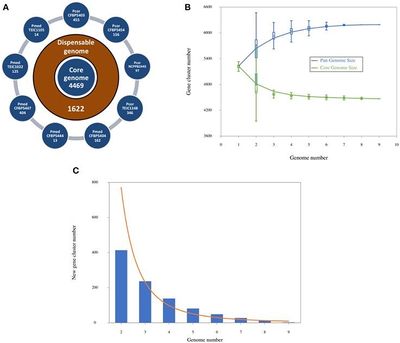
Genome size is approximately 6,193,893, the DNA Coding Region has 4,577,116 base pairs, DNA G+C content 60.51%, Total Genes 6,211, Protein Coding Genes 6,165, Genes with Predicted Function 4,074. (Licciardello, et al, 2014)
Cell structure and metabolism
P. corrugata is a gram-negative, oxidase-positive, non-spore forming rod, and non-fluorescent on King's B medium (Catara, et al, 2007). P. corrugata produces wrinkled and, rarely, smooth colonies on yeast peptone glucose agar or nutrient dextrose agar; yellow to brown diffusible pigments are frequently produced. Disease symptoms include: typical symptom on tomato is necrosis and/or hollowing of the pith of the stem, the syndrome determines loss of turgidity of the plant, hydropic/necrotic areas, and long conspicuous adventitious roots on the stem. Also, there is no production of fluorescent compounds, it builds up medium sized chains of poly(hydroxyalkanoates), and is incapable of producing levan (Catara, et al, 2002).
Ecology and Pathogenesis
Plant-pathogenic bacterium that causes pith necrosis in tomatoes (Anzai, et al, 2000). The first case was reported in April of 2015 in two countries simultaneously in Georgia (Kůdela. Krejzar, and Pánková, 2010). A range of 10% to 15% of the crops were affected (Kůdela, et al, 2010). Infections are more evident when the first trusses ripen on the tomato plants. Initially, only slight wilting in the hottest part of the day and at times chlorosis of the apex can be seen. The characteristic symptom of the disease, as shown by its name, is stem pith necrosis and the pith may appear as: necrotic, dry and slightly disaggregated in the core; hydropic, white or dark but presenting a hard core and necrotic in the areas near the xylom. The disease starts from the base of the stem and works up to the leaf stems and bunches. Necrosis can also affect the taproot and, occasionally, the rootlets (Catara, Sutra, et al., 2002). Under test conditions 'Pseudomonas corrugata' was also able to infect peppers, tobacco, and chrysanthemum (Catara 2002).
Current Research
Caption: Current research involving P. corrugata includes: ● “Association with the collapse of tomato plants in rockwool slab hydroponic culture” (Kůdela V. et. al, 2010). This study is attempting to understand how pith occurs in tomato plants that could trigger an outbreak such as the Rockwool Slab Hydroponic incident in 2008. ● Suppression of damping-off in maize seedlings by P. corrugata (Pandey et. al., 2001). This is a method that may be able to improve grain yields by introducing it to corn seedlings. ● Pseudomonas corrugata (NRRL B-30409) Mutants Increased Phosphate Solubilization, Organic Acid Production, and Plant Growth at Lower Temperatures (Trivedi et. al., 2007). This is again another agricultural method that researchers are hoping will improve crop yields.
References
1. Anzai; Kim, H; Park, JY; Wakabayashi, H; Oyaizu, H; et al. (Jul 2000). "Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence". Int J Syst Evol Microbiol. 50 (4): 1563–89. doi:10.1099/00207713-50-4-1563. PMID 10939664. 2. Catara, Vittoria (2007). "Pseudomonas Corrugata: Plant Pathogen And/or Biological Resource?" Molecular Plant Pathology 8.3: 233-44. Wiley Online Library. 3. Catara, Vittoria, et al (2002). "Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov." International Journal of Systematic and Evolutionary Microbiology 52.5: 1749-1758. 4. Kůdela, V., V. Krejzar, and L. Pánková (2010). "Pseudomonas Corrugata and Pseudomonas Marginalis Associated with the Collapse of Tomato Plants in Rockwool Slab Hydroponic Culture." Plant Protection Science 46.1: 1-11. 5. Licciardello, G., Ferraro, R., Russo, M., Strozzi, F., Catara, A. F., Bella, P., & Catara, V. (2016). Transcriptome analysis of Pseudomonas mediterranea and P. corrugata plant pathogens during accumulation of medium-chain-length PHAs by glycerol bioconversion. New Biotechnology. 6. Licciardello, G., et al. "Draft genome sequence of Pseudomonas corrugata, a phytopathogenic bacterium with potential industrial applications." Journal of biotechnology 175 (2014): 65-66. 7. Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A. P., ... & Brinkac, L. (2002). Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environmental microbiology, 4(12), 799-808. 8. Pandey, Anita, Lok Man S. Palni, and K.p. Hebbar (2001). "Suppression of Damping-off in Maize Seedlings by Pseudomonas Corrugata." Microbiological Research 156.2: 191-94. 9. Pandey, Anita, Lok Man S.Palni, and Pankaj, Trivedi (2008). “In vitro evaluation of antagonistic properties of Pseudomonas corrugata”. Microbiological Research. 10. Smith, Dunez, Lelliot, Phillips and Archer (1988). “European Handbook of Plant Disease”. Blackwell Scientific Publications. 11. Trantas; Sarris, PF; Pentari, MG; Mpalantinaki, E; Ververidis, F; Goumas, DE; et al. "Diversity among Pseudomonas corrugata and Pseudomonas mediterranea isolated from tomato and pepper showing symptoms of pith necrosis in Greece". Plant Pathology. 64 (2): 307–318. doi:10.1111/ppa.12261 12. Trantas, Emmanouil A., Grazia Licciardello, Nalvo F. Almeida, Kamil Witek, Cinzia P. Strano, Zane Duxbury, Filippos Ververidis, Dimitrios E. Goumas, Jonathan D. G. Jones, David S. Guttman, Vittoria Catara, and Panagiotis F. Sarris. (2015). "Comparative Genomic Analysis of Multiple Strains of Two Unusual Plant Pathogens: Pseudomonas Corrugata and Pseudomonas Mediterranea." Front. Microbiol. Frontiers in Microbiology 6.811: 1-19. ResearchGate. 13. Trantas, Emmanouil A., et al. "Comparative genomic analysis of multiple strains of two unusual plant pathogens: Pseudomonas corrugata and Pseudomonas mediterranea." Frontiers in microbiology 6 (2015). 14. Trivedi, Pankaj, and Tongmin Sa (2007). "Pseudomonas Corrugata (NRRL B-30409) Mutants Increased Phosphate Solubilization, Organic Acid Production, and Plant Growth at Lower Temperatures." Current Microbiology 56.2: 140-44.
9. Author: Cameron Cole, Alex Roman, and Lexi Siegle with Microbial Ecology Instructor Dr. Hideotoshi Urakawa