Reptile-Exotic-Pet-Associated-Salmonellosis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
<br>By Tomas Grant<br> | <br>By Tomas Grant<br> | ||
==Introduction== | ==Introduction== | ||
<br>Salmonellosis is one of the most common foodbourne diseases and can be caused by the various serovars of the species <i>S. enterica </i>, and is found worldwide, causing 93.8 million cases of gastroenteritis and over 155,00 deaths annually worldwide (Noellie et el 2014). Understanding the pathology of foodbourne Salmonellosis is important however, there is an increasing concern for Reptile-Exotic-Pet-Associated-Salmonellosis (REPAS) , a major agent of infection through direct or indirect animal contact in people’s homes, veterinary clinics, farms, zoological gardens, and other professional, public and private settings. (Rabsch et, al, “The Zoonotic agent Salmonellosis) . There are a number of reports that describe the prevalence of <i>Salmonella</i> spp. in reptiles ranging from lizards, snakes, and turtles, and multiple serovars have been identified and shown to be associated with each of these organisms (Sylvester et al, 2014). Reptiles have been shown to be asymptomatic carriers of <i>Salmonella</i> and have been shown to have a natural interaction with the bacteria, with these bacteria being natural inhabitants of reptile gut microflora, shed at various rates throughout the reptile’s lifecycle. Reptiles are often unrecognized sources for the disease in humans and there are estimates that indicate reptile-associated salmonellosis is responsible for 3-11% of all human salmonellosis cases within both Canada and the United States (Rabsch et al, 2014). | |||
<i>Salmonella</i> spp. are rod-shaped, flagellated, and facultative anaerobic bacteria which belong to the family Enterobacteriaceae, and as apart of the phylum Proteobacteria are Gram-negative. Within the genus <i>Salmonella</i> there are two distinct species; <i>S. enterica</i> and <I> S. bongori </i>, but the majority of the diversity lies within the more than 2,600 servovars of the species <i>S. enterica </i>. The three main serovars within this species are (1) <i>S. typhi</i>, the cause of systemic infections and typhoid fever, (2) <i>S. Enteritidis</i>, a major food cause of food poisoning associated with poultry farming, and (3) <i>S. typhimurium</i>, which is not fatal in humans, but may cause gastroenteritis, but also more serious cases including septicemia, meningitis, and subnormal empyema (Rabsch et, al, 2014). | |||
[[Image:fig142x.jpg|thumb|350px|right|''Salmonella''. Courtesy of [http://www.meddean.luc.edu/lumen/MedEd/orfpath/images/fig135x.jpg LUMEN.]]] | [[Image:fig142x.jpg|thumb|350px|right|''Salmonella''. Courtesy of [http://www.meddean.luc.edu/lumen/MedEd/orfpath/images/fig135x.jpg LUMEN.]]] | ||
<br>P2</br> | <br>P2</br> | ||
Revision as of 17:35, 22 April 2015
By Tomas Grant
Introduction
Salmonellosis is one of the most common foodbourne diseases and can be caused by the various serovars of the species S. enterica , and is found worldwide, causing 93.8 million cases of gastroenteritis and over 155,00 deaths annually worldwide (Noellie et el 2014). Understanding the pathology of foodbourne Salmonellosis is important however, there is an increasing concern for Reptile-Exotic-Pet-Associated-Salmonellosis (REPAS) , a major agent of infection through direct or indirect animal contact in people’s homes, veterinary clinics, farms, zoological gardens, and other professional, public and private settings. (Rabsch et, al, “The Zoonotic agent Salmonellosis) . There are a number of reports that describe the prevalence of Salmonella spp. in reptiles ranging from lizards, snakes, and turtles, and multiple serovars have been identified and shown to be associated with each of these organisms (Sylvester et al, 2014). Reptiles have been shown to be asymptomatic carriers of Salmonella and have been shown to have a natural interaction with the bacteria, with these bacteria being natural inhabitants of reptile gut microflora, shed at various rates throughout the reptile’s lifecycle. Reptiles are often unrecognized sources for the disease in humans and there are estimates that indicate reptile-associated salmonellosis is responsible for 3-11% of all human salmonellosis cases within both Canada and the United States (Rabsch et al, 2014).
Salmonella spp. are rod-shaped, flagellated, and facultative anaerobic bacteria which belong to the family Enterobacteriaceae, and as apart of the phylum Proteobacteria are Gram-negative. Within the genus Salmonella there are two distinct species; S. enterica and S. bongori , but the majority of the diversity lies within the more than 2,600 servovars of the species S. enterica . The three main serovars within this species are (1) S. typhi, the cause of systemic infections and typhoid fever, (2) S. Enteritidis, a major food cause of food poisoning associated with poultry farming, and (3) S. typhimurium, which is not fatal in humans, but may cause gastroenteritis, but also more serious cases including septicemia, meningitis, and subnormal empyema (Rabsch et, al, 2014).
P2
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Classification
Higher order taxa:
Species:
Salmonella enterica subsp. I serovar Typhimurium (S. typhimurium LT2), S. enterica subsp. enterica serovar Typhi (S. typhi CT18), S. enterica subsp. enterica serovar Typhi Ty2 (S. typhi Ty2)
NCBI: Taxonomy Genome: S. typhi CT18 S. typhi Ty2 S. typhimurium|}Epidemiology
Transmission of Salmonella
P1
P2
Pathology and Immune Response
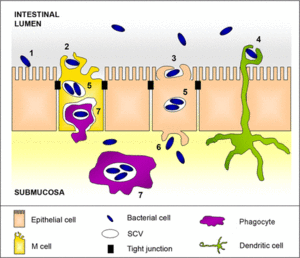
Clinical Signs of Infection
Pathogenesis of Salmonella serovar S. Typhimurium
P1
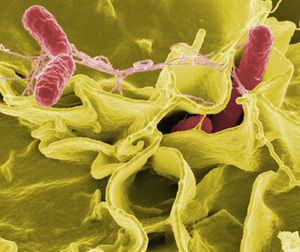
P2
P3
P4
Antibiotic Resistance and Treatment
Risks Associated With S. enterica infections
Risk Factors

Disease Prevention Measures
References
[1]
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2015, Kenyon College.
