Reptile-Exotic-Pet-Associated-Salmonellosis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
<br>By Tomas Grant<br> | <br>By Tomas Grant<br> | ||
==Introduction== | ==Introduction== | ||
<br>Salmonellosis is one of the most common foodbourne diseases and can be caused by the various serovars of the species <i>S. enterica </i>, and is found worldwide, causing 93.8 million cases of gastroenteritis and over 155,00 deaths annually worldwide (Noellie et el 2014). Understanding the pathology of foodbourne Salmonellosis is important however, there is an increasing concern for Reptile-Exotic-Pet-Associated-Salmonellosis (REPAS) , a major agent of infection through direct or indirect animal contact in people’s homes, veterinary clinics, farms, zoological gardens, and other professional, public and private settings. (Rabsch et, al, “The Zoonotic agent Salmonellosis) . There are a number of reports that describe the prevalence of <i>Salmonella</i> spp. in reptiles ranging from lizards, snakes, and turtles, and multiple serovars have been identified and shown to be associated with each of these organisms (Sylvester et al, 2014). Reptiles have been shown to be asymptomatic carriers of <i>Salmonella</i> and have been shown to have a natural interaction with the bacteria, with these bacteria being natural inhabitants of reptile gut microflora, shed at various rates throughout the reptile’s lifecycle. Reptiles are often unrecognized sources for the disease in humans and there are estimates that indicate reptile-associated salmonellosis is responsible for 3-11% of all human salmonellosis cases within both Canada and the United States (Rabsch et al, 2014). | <br>Salmonellosis is one of the most common foodbourne diseases and can be caused by the various serovars of the species <i>S. enterica </i>, and is found worldwide, causing 93.8 million cases of gastroenteritis and over 155,00 deaths annually worldwide (Noellie et el 2014). Understanding the pathology of foodbourne Salmonellosis is important however, there is an increasing concern for Reptile-Exotic-Pet-Associated-Salmonellosis (REPAS) , a major agent of infection through direct or indirect animal contact in people’s homes, veterinary clinics, farms, zoological gardens, and other professional, public and private settings. (Rabsch et, al, “The Zoonotic agent Salmonellosis). There are a number of reports that describe the prevalence of <i>Salmonella</i> spp. in reptiles ranging from lizards, snakes, and turtles, and multiple serovars have been identified and shown to be associated with each of these organisms (Sylvester et al, 2014). Reptiles have been shown to be asymptomatic carriers of <i>Salmonella</i> and have been shown to have a natural interaction with the bacteria, with these bacteria being natural inhabitants of reptile gut microflora, shed at various rates throughout the reptile’s lifecycle. Reptiles are often unrecognized sources for the disease in humans and there are estimates that indicate reptile-associated salmonellosis is responsible for 3-11% of all human salmonellosis cases within both Canada and the United States (Rabsch et al, 2014). | ||
<i>Salmonella</i> spp. are rod-shaped, flagellated, and facultative anaerobic bacteria which belong to the family Enterobacteriaceae, and as apart of the phylum Proteobacteria are Gram-negative. Within the genus <i>Salmonella</i> there are two distinct species; <i>S. enterica</i> and <I> S. bongori </i>, but the majority of the diversity lies within the more than 2,600 servovars of the species <i>S. enterica </i>. The three main serovars within this species are (1) <i>S. typhi</i>, the cause of systemic infections and typhoid fever, (2) <i>S. Enteritidis</i>, a major food cause of food poisoning associated with poultry farming, and (3) <i>S. typhimurium</i>, which is not fatal in humans, but may cause gastroenteritis, but also more serious cases including septicemia, meningitis, and subnormal empyema (Rabsch et, al, 2014). | <i>Salmonella</i> spp. are rod-shaped, flagellated, and facultative anaerobic bacteria which belong to the family Enterobacteriaceae, and as apart of the phylum Proteobacteria are Gram-negative. Within the genus <i>Salmonella</i> there are two distinct species; <i>S. enterica</i> and <I> S. bongori </i>, but the majority of the diversity lies within the more than 2,600 servovars of the species <i>S. enterica </i>. The three main serovars within this species are (1) <i>S. typhi</i>, the cause of systemic infections and typhoid fever, (2) <i>S. Enteritidis</i>, a major food cause of food poisoning associated with poultry farming, and (3) <i>S. typhimurium</i>, which is not fatal in humans, but may cause gastroenteritis, but also more serious cases including septicemia, meningitis, and subnormal empyema (Rabsch et, al, 2014). | ||
| Line 7: | Line 7: | ||
[[Image:fig142x.jpg|thumb|350px|right|''Salmonella''. Courtesy of [http://www.meddean.luc.edu/lumen/MedEd/orfpath/images/fig135x.jpg LUMEN.]]] | [[Image:fig142x.jpg|thumb|350px|right|''Salmonella''. Courtesy of [http://www.meddean.luc.edu/lumen/MedEd/orfpath/images/fig135x.jpg LUMEN.]]] | ||
<br>P2</br> | <br>P2</br> | ||
==Classification== | ==Classification== | ||
| Line 34: | Line 20: | ||
==Epidemiology== | ==Epidemiology== | ||
===Transmission of Salmonella=== | ===Transmission of Salmonella=== | ||
The main pathway of transmission for <i>Salmonella enterica</i> spp., which is an invasive primary pathogen, is by the fecal-oral route which usually begins with the ingestion of contaminated water or food, so that the bacterium is able to encounter the intestinal epithelium, spreading throughout the gastrointestinal tract causing gastroenteritis. Humans typically acquire the organisms through either direct transmission by handling a reptile, and via indirect transmission by coming into contact with any object that has been contaminated by a reptile. Other sources of transmission include aquariums, terrariums, cages, as well as the water that most reptiles like turtles and amphibians live in. Clothing that has come into direct contact with reptiles has been associated as another source of transmission from one infected individual, and also bites and claw scratches have been shown to be effective at transmitting <i>Salmonella</i> spp. | |||
<br> | |||
<i>Salmonella</i> has ben shown to be able to survive for long periods of time in the environment and prefers those environments where it is humid and warm, and has been isolated for prolonged periods from a multitude of surfaces that have been contaminated by reptile fecal matter (Centers for Disease Control, Jan 2013). It is of important note to highlight the fact that the CDC found that <i>Salmonella</i> has been reported to “survive 115 days in pond water, 89 days in tap water, within dried reptile feces, and from aquariums and terrarium water up to 6 weeks after removal of a turtle from the habitat (CDC, Jan 2013). The disease can be transmitted to other individuals, both human and animal alike in fecal matter, with the organisms being shed continually throughout the duration of the infection. Shedding has been shown to last anywhere from several days to multiple weeks, with the ability of certain individuals to become asymptomatic carriers for several months. It has been shown that non-typhoidal Salmonellosis infection patients have the ability to shed the bacteria in their feces for over a year, and antibiotic treatment has been shown to typically prolong the shedding of these organisms during this time period (CDC, Jan 2013). Also, over 90% of reptiles may carry <i>Salmonella</i> and these animals may carry up to five different serotypes, increasing the risk associated with reptiles as a whole. Also,it is important to note that recent studies have shown that shedding of <i>Salmonella</i> within the feces is higher in reptiles that are kept in captivity when compared to wild reptiles (Rabsch et al), which is especially important because these “pet” reptiles are kept in closer proximity and are in closer contact to humans. These findings alone show the danger associated with such a long living organism and help demonstrate the prevalence of the disease Salmonellosis and the dangers associated with <i>Salmonella</i> as a world-wide issue. | |||
<br><i>Salmonella</i> has ben shown to be able to survive for long periods of time in the environment and prefers those environments where it is humid and warm, and has been isolated for prolonged periods from a multitude of surfaces that have been contaminated by reptile fecal matter (Centers for Disease Control, Jan 2013). It is of important note to highlight the fact that the CDC found that <i>Salmonella</i> has been reported to “survive 115 days in pond water, 89 days in tap water, within dried reptile feces, and from aquariums and terrarium water up to 6 weeks after removal of a turtle from the habitat (CDC, Jan 2013). The disease can be transmitted to other individuals, both human and animal alike in fecal matter, with the organisms being shed continually throughout the duration of the infection. Shedding has been shown to last anywhere from several days to multiple weeks, with the ability of certain individuals to become asymptomatic carriers for several months. It has been shown that non-typhoidal Salmonellosis infection patients have the ability to shed the bacteria in their feces for over a year, and antibiotic treatment has been shown to typically prolong the shedding of these organisms during this time period (CDC, Jan 2013). Also, over 90% of reptiles may carry <i>Salmonella</i> and these animals may carry up to five different serotypes, increasing the risk associated with reptiles as a whole. Also,it is important to note that recent studies have shown that shedding of <i>Salmonella</i> within the feces is higher in reptiles that are kept in captivity when compared to wild reptiles (Rabsch et al), which is especially important because these “pet” reptiles are kept in closer proximity and are in closer contact to humans. These findings alone show the danger associated with such a long living organism and help demonstrate the prevalence of the disease Salmonellosis and the dangers associated with <i>Salmonella</i> as a world-wide issue. </br> | |||
===Pathology and Immune Response=== | ===Pathology and Immune Response=== | ||
====Clinical Signs of Infection==== | |||
Patients infected by <i>Salmonella</i> typically present a multitude of symptoms including an acute onset of fever, abdominal pain, cramping, as well as diarrhea with or without blood (typically associated with inflammation of the large bowel), as well as vomiting and severe nausea, but there is a wide spectrum of severity of the illness which may be indicated by only a few or a multitude of symptoms (Fabrega and Vila, 2013). The disease usually manifests after the ingestion of >45,000 bacteria in contaminated food, water, or other sources of infection and non-typhoidal, self-limiting, <i>Salmonella</i> gastroenteritis requires a period of incubation ranging from 6 to 72 hours depending on susceptibility of the host (CDC Jan 2013). Whether or not <i>Salmonella</i> remains within the intestine or disseminates to other major organs depends mainly on the host’s immune response as well as the virulence factors the strain possesses. | |||
====Pathogenesis of <i>Salmonella</i> serovar <i> S. Typhimurium </i>==== | |||
The pathogenesis triggered by the <i>Salmonella</i> serovar <i> S. Typhimurium <i/> has been studied extensively and knowledge about its multitude of virulence mechanisms for successfully invading host tissue cells has been compiled. These organisms have a large armamentarium of virulence factors all regulated by an extremely complex regulatory meshwork, responsible for coordinating and synchronizing all the time sensitive elements involved. This regulatory mechanism is extremely important because it guarantees expression of each virulence element, but it also facilitates a communication between all of the determinants, ensuring the invading bacteria responds appropriately at all stages in a temporal hierarchal method. These elements include <i>Salmonella</i> pathogenicity islands (SPI’s), as well as other virulence determinants, including those encoded within the pSLT virulence plasmid, as well as adhesions, flagella, and biofilm forming proteins which all contribute to several stages of the disease (Fabrega and Vila, 2013). | |||
[[Image:TKGF3.medium.gif|thumb|300px|left|<b>Figure 1-Salmonella enterica Typhimurium pathology model within the human intestine.The bacteria begins the process with attachment to intestinal epithelium (1) via adhesins, followed by invasion, and then engulfment of bacteria (2,3). Salmonella is localized inside the SVC once inside the cytoplasm, where it replicates (5). SVC's trancytose these cells and enter the basolateral membrane, and then the cells within the SVC are released to the submucosa (6) where they become internalized within recruited phagocytes (7), followed by subsequent dissemination by the bloodstream.]]] | [[Image:TKGF3.medium.gif|thumb|300px|left|<b>Figure 1-Salmonella enterica Typhimurium pathology model within the human intestine.The bacteria begins the process with attachment to intestinal epithelium (1) via adhesins, followed by invasion, and then engulfment of bacteria (2,3). Salmonella is localized inside the SVC once inside the cytoplasm, where it replicates (5). SVC's trancytose these cells and enter the basolateral membrane, and then the cells within the SVC are released to the submucosa (6) where they become internalized within recruited phagocytes (7), followed by subsequent dissemination by the bloodstream.]]] | ||
The first step in the infection pathway of <i>Salmonella</i> Typhimurium involves host ingestion of <i>Salmonella</i> found in contaminated food or water as mentioned before, and the subsequent invasion of the stomach which presents the first obstacle to successful infection: the highly acidic pH of the host organism’s stomach. In order to overcome this obstacle, <i>S</i>. Typhimurium activates the acid tolerance response (ATR) system, which provides an inducible pH-homeostatic function in order to maintain the organism’s intracellular pH at levels higher than those found in the stomach (Fabrega and Vila, 2013). Upon entry into the small intestine the organism must adhere to and then cross the intestinal mucus layer, the second line of defense against pathogens, and then must adhere to intestinal epithelial cells, causing the appearance of the infection in the form of large cytoskeletal rearrangements within host tissue cells. An important feature of these bacteria are the presence of flagella which gives <i>Salmonella</i> an increased chance at encountering the intestinal epithelium, leading to subsequent adhesion and invasion of host cell tissues. The intestinal epithelium is the primary interface between the internal and external environment, and the adhesion of <i>Salmonella</i> is a key factor in its pathogenicity, or its ability to cause disease. Approach and attachment are enhanced by the motility of the organism, which are driven by several virulence factors including multiple adhesins and fimbriae. | |||
[[Image:yellow_salmonella.JPG|thumb|300px|right|''Salmonella typhimurium'' (red) invading cultured human cells. Courtesy of [http://www.asknature.org/strategy/2312b4c3ecfc07d04ada7c75ea9fd3cc#.VTVExq1Viko Ask Nature]]] | [[Image:yellow_salmonella.JPG|thumb|300px|right|''Salmonella typhimurium'' (red) invading cultured human cells. Courtesy of [http://www.asknature.org/strategy/2312b4c3ecfc07d04ada7c75ea9fd3cc#.VTVExq1Viko Ask Nature]]] | ||
<br> | <br>The internal modifications induced within host cells serves to disrupt normal epithelial brush borders and induces the formation of membrane ruffles. These ruffles are responsible for engulfing the adhered bacterium in large vesicles called <i>Salmonella</i>-containing vacuoles (SCVs) (Fabrega and Vila, 2013). Engulfment of <i>Salmonella</i> is mediated by virulence elements encoded within SP1-I, which encodes for several effector proteins responsible for triggering the invasion of epithelial cells within the membrane, and regulates actin cytoskeletal rearrangements which leads to the intake of these bacteria (Figure1- 2,3,5). (Fabrega and Vila, 2013) These SVC’s are the most important determinants of <i>Salmonella</i> pathogenicity because they represent the intracellular compartments in which these organisms can replicate,and highlights the facultative intracellular nature of these bacteria. SVC’s arise during the early endocytic pathway, but must direct changes within host endocytic trafficking and function to avoid direct fusion of SVC’s with secondary lysomsomes and halt activity of lysosomal enzymes. An important mechanism for the maintenance of the integrity of the SVC membrane is the <i>Salmonella</i> induced formation of an F-actin meshwork which surrounds bacterial vacuoles, a process known as vacuole-associated actin polymerization (VAP) (Fabrega et al, 2013). This not only protects the bacterial SVC’s, but allows for the migration of these vacuoles to the perinuclear position near the Golgi apparatus, which allows for the interception of host exo- and endo-cytic transport vesicles to obtain nutrients essential for supporting bacterial replication (Fabrega et al, 2013). | ||
While initially forming the SVC’s, the invading <i>Salmonella</i> bacterium induces a secretory response within the intestinal epithelium that aims to recruit and transmigrate phagocytes located in the submucosal space across the membrane and into intestinal lumen, followed by the reconstitution of the once disrupted epithelial brush border. Those phagocytes now in the intestinal lumen can be used to engulf individual cells and transport them directly across the membrane. A number of SVC’s also migrate to the basolateral membrane, release their internal cells into the submucosa, and then engulfed by phagocytes. More specifically three phagocytic types have been reported to interact directly with invading <i>Salmonella</i> bacteria including (i) neutrophils, (ii) inflammatory monocytes, and (iii) dendritic cells (Fig1-6 and 7) (Fabrega and Vila, 2013). These bacteria are then internalized again within an SCV allowing for further replication, and the migration of infected phagocytes allows for the spread of the invading bacteria, and subsequent circulation in the host’s bloodstream to several tissues including the spleen, liver, and in extreme cases the brain. | |||
<br> | <br> | ||
==Antibiotic Resistance and Treatment== | ==Antibiotic Resistance and Treatment== | ||
Revision as of 17:39, 22 April 2015
By Tomas Grant
Introduction
Salmonellosis is one of the most common foodbourne diseases and can be caused by the various serovars of the species S. enterica , and is found worldwide, causing 93.8 million cases of gastroenteritis and over 155,00 deaths annually worldwide (Noellie et el 2014). Understanding the pathology of foodbourne Salmonellosis is important however, there is an increasing concern for Reptile-Exotic-Pet-Associated-Salmonellosis (REPAS) , a major agent of infection through direct or indirect animal contact in people’s homes, veterinary clinics, farms, zoological gardens, and other professional, public and private settings. (Rabsch et, al, “The Zoonotic agent Salmonellosis). There are a number of reports that describe the prevalence of Salmonella spp. in reptiles ranging from lizards, snakes, and turtles, and multiple serovars have been identified and shown to be associated with each of these organisms (Sylvester et al, 2014). Reptiles have been shown to be asymptomatic carriers of Salmonella and have been shown to have a natural interaction with the bacteria, with these bacteria being natural inhabitants of reptile gut microflora, shed at various rates throughout the reptile’s lifecycle. Reptiles are often unrecognized sources for the disease in humans and there are estimates that indicate reptile-associated salmonellosis is responsible for 3-11% of all human salmonellosis cases within both Canada and the United States (Rabsch et al, 2014).
Salmonella spp. are rod-shaped, flagellated, and facultative anaerobic bacteria which belong to the family Enterobacteriaceae, and as apart of the phylum Proteobacteria are Gram-negative. Within the genus Salmonella there are two distinct species; S. enterica and S. bongori , but the majority of the diversity lies within the more than 2,600 servovars of the species S. enterica . The three main serovars within this species are (1) S. typhi, the cause of systemic infections and typhoid fever, (2) S. Enteritidis, a major food cause of food poisoning associated with poultry farming, and (3) S. typhimurium, which is not fatal in humans, but may cause gastroenteritis, but also more serious cases including septicemia, meningitis, and subnormal empyema (Rabsch et, al, 2014).
P2
Classification
Higher order taxa:
Species:
Salmonella enterica subsp. I serovar Typhimurium (S. typhimurium LT2), S. enterica subsp. enterica serovar Typhi (S. typhi CT18), S. enterica subsp. enterica serovar Typhi Ty2 (S. typhi Ty2)
NCBI: Taxonomy Genome: S. typhi CT18 S. typhi Ty2 S. typhimurium|}Epidemiology
Transmission of Salmonella
The main pathway of transmission for Salmonella enterica spp., which is an invasive primary pathogen, is by the fecal-oral route which usually begins with the ingestion of contaminated water or food, so that the bacterium is able to encounter the intestinal epithelium, spreading throughout the gastrointestinal tract causing gastroenteritis. Humans typically acquire the organisms through either direct transmission by handling a reptile, and via indirect transmission by coming into contact with any object that has been contaminated by a reptile. Other sources of transmission include aquariums, terrariums, cages, as well as the water that most reptiles like turtles and amphibians live in. Clothing that has come into direct contact with reptiles has been associated as another source of transmission from one infected individual, and also bites and claw scratches have been shown to be effective at transmitting Salmonella spp.
Salmonella has ben shown to be able to survive for long periods of time in the environment and prefers those environments where it is humid and warm, and has been isolated for prolonged periods from a multitude of surfaces that have been contaminated by reptile fecal matter (Centers for Disease Control, Jan 2013). It is of important note to highlight the fact that the CDC found that Salmonella has been reported to “survive 115 days in pond water, 89 days in tap water, within dried reptile feces, and from aquariums and terrarium water up to 6 weeks after removal of a turtle from the habitat (CDC, Jan 2013). The disease can be transmitted to other individuals, both human and animal alike in fecal matter, with the organisms being shed continually throughout the duration of the infection. Shedding has been shown to last anywhere from several days to multiple weeks, with the ability of certain individuals to become asymptomatic carriers for several months. It has been shown that non-typhoidal Salmonellosis infection patients have the ability to shed the bacteria in their feces for over a year, and antibiotic treatment has been shown to typically prolong the shedding of these organisms during this time period (CDC, Jan 2013). Also, over 90% of reptiles may carry Salmonella and these animals may carry up to five different serotypes, increasing the risk associated with reptiles as a whole. Also,it is important to note that recent studies have shown that shedding of Salmonella within the feces is higher in reptiles that are kept in captivity when compared to wild reptiles (Rabsch et al), which is especially important because these “pet” reptiles are kept in closer proximity and are in closer contact to humans. These findings alone show the danger associated with such a long living organism and help demonstrate the prevalence of the disease Salmonellosis and the dangers associated with Salmonella as a world-wide issue.
Salmonella has ben shown to be able to survive for long periods of time in the environment and prefers those environments where it is humid and warm, and has been isolated for prolonged periods from a multitude of surfaces that have been contaminated by reptile fecal matter (Centers for Disease Control, Jan 2013). It is of important note to highlight the fact that the CDC found that Salmonella has been reported to “survive 115 days in pond water, 89 days in tap water, within dried reptile feces, and from aquariums and terrarium water up to 6 weeks after removal of a turtle from the habitat (CDC, Jan 2013). The disease can be transmitted to other individuals, both human and animal alike in fecal matter, with the organisms being shed continually throughout the duration of the infection. Shedding has been shown to last anywhere from several days to multiple weeks, with the ability of certain individuals to become asymptomatic carriers for several months. It has been shown that non-typhoidal Salmonellosis infection patients have the ability to shed the bacteria in their feces for over a year, and antibiotic treatment has been shown to typically prolong the shedding of these organisms during this time period (CDC, Jan 2013). Also, over 90% of reptiles may carry Salmonella and these animals may carry up to five different serotypes, increasing the risk associated with reptiles as a whole. Also,it is important to note that recent studies have shown that shedding of Salmonella within the feces is higher in reptiles that are kept in captivity when compared to wild reptiles (Rabsch et al), which is especially important because these “pet” reptiles are kept in closer proximity and are in closer contact to humans. These findings alone show the danger associated with such a long living organism and help demonstrate the prevalence of the disease Salmonellosis and the dangers associated with Salmonella as a world-wide issue.
Pathology and Immune Response
Clinical Signs of Infection
Patients infected by Salmonella typically present a multitude of symptoms including an acute onset of fever, abdominal pain, cramping, as well as diarrhea with or without blood (typically associated with inflammation of the large bowel), as well as vomiting and severe nausea, but there is a wide spectrum of severity of the illness which may be indicated by only a few or a multitude of symptoms (Fabrega and Vila, 2013). The disease usually manifests after the ingestion of >45,000 bacteria in contaminated food, water, or other sources of infection and non-typhoidal, self-limiting, Salmonella gastroenteritis requires a period of incubation ranging from 6 to 72 hours depending on susceptibility of the host (CDC Jan 2013). Whether or not Salmonella remains within the intestine or disseminates to other major organs depends mainly on the host’s immune response as well as the virulence factors the strain possesses.
Pathogenesis of Salmonella serovar S. Typhimurium
The pathogenesis triggered by the Salmonella serovar S. Typhimurium has been studied extensively and knowledge about its multitude of virulence mechanisms for successfully invading host tissue cells has been compiled. These organisms have a large armamentarium of virulence factors all regulated by an extremely complex regulatory meshwork, responsible for coordinating and synchronizing all the time sensitive elements involved. This regulatory mechanism is extremely important because it guarantees expression of each virulence element, but it also facilitates a communication between all of the determinants, ensuring the invading bacteria responds appropriately at all stages in a temporal hierarchal method. These elements include Salmonella pathogenicity islands (SPI’s), as well as other virulence determinants, including those encoded within the pSLT virulence plasmid, as well as adhesions, flagella, and biofilm forming proteins which all contribute to several stages of the disease (Fabrega and Vila, 2013).
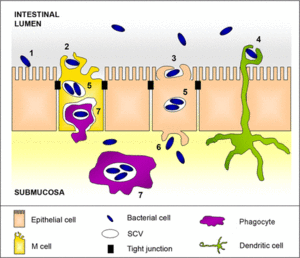
The first step in the infection pathway of Salmonella Typhimurium involves host ingestion of Salmonella found in contaminated food or water as mentioned before, and the subsequent invasion of the stomach which presents the first obstacle to successful infection: the highly acidic pH of the host organism’s stomach. In order to overcome this obstacle, S. Typhimurium activates the acid tolerance response (ATR) system, which provides an inducible pH-homeostatic function in order to maintain the organism’s intracellular pH at levels higher than those found in the stomach (Fabrega and Vila, 2013). Upon entry into the small intestine the organism must adhere to and then cross the intestinal mucus layer, the second line of defense against pathogens, and then must adhere to intestinal epithelial cells, causing the appearance of the infection in the form of large cytoskeletal rearrangements within host tissue cells. An important feature of these bacteria are the presence of flagella which gives Salmonella an increased chance at encountering the intestinal epithelium, leading to subsequent adhesion and invasion of host cell tissues. The intestinal epithelium is the primary interface between the internal and external environment, and the adhesion of Salmonella is a key factor in its pathogenicity, or its ability to cause disease. Approach and attachment are enhanced by the motility of the organism, which are driven by several virulence factors including multiple adhesins and fimbriae.
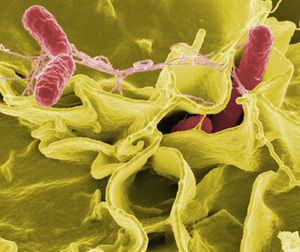
The internal modifications induced within host cells serves to disrupt normal epithelial brush borders and induces the formation of membrane ruffles. These ruffles are responsible for engulfing the adhered bacterium in large vesicles called Salmonella-containing vacuoles (SCVs) (Fabrega and Vila, 2013). Engulfment of Salmonella is mediated by virulence elements encoded within SP1-I, which encodes for several effector proteins responsible for triggering the invasion of epithelial cells within the membrane, and regulates actin cytoskeletal rearrangements which leads to the intake of these bacteria (Figure1- 2,3,5). (Fabrega and Vila, 2013) These SVC’s are the most important determinants of Salmonella pathogenicity because they represent the intracellular compartments in which these organisms can replicate,and highlights the facultative intracellular nature of these bacteria. SVC’s arise during the early endocytic pathway, but must direct changes within host endocytic trafficking and function to avoid direct fusion of SVC’s with secondary lysomsomes and halt activity of lysosomal enzymes. An important mechanism for the maintenance of the integrity of the SVC membrane is the Salmonella induced formation of an F-actin meshwork which surrounds bacterial vacuoles, a process known as vacuole-associated actin polymerization (VAP) (Fabrega et al, 2013). This not only protects the bacterial SVC’s, but allows for the migration of these vacuoles to the perinuclear position near the Golgi apparatus, which allows for the interception of host exo- and endo-cytic transport vesicles to obtain nutrients essential for supporting bacterial replication (Fabrega et al, 2013).
While initially forming the SVC’s, the invading Salmonella bacterium induces a secretory response within the intestinal epithelium that aims to recruit and transmigrate phagocytes located in the submucosal space across the membrane and into intestinal lumen, followed by the reconstitution of the once disrupted epithelial brush border. Those phagocytes now in the intestinal lumen can be used to engulf individual cells and transport them directly across the membrane. A number of SVC’s also migrate to the basolateral membrane, release their internal cells into the submucosa, and then engulfed by phagocytes. More specifically three phagocytic types have been reported to interact directly with invading Salmonella bacteria including (i) neutrophils, (ii) inflammatory monocytes, and (iii) dendritic cells (Fig1-6 and 7) (Fabrega and Vila, 2013). These bacteria are then internalized again within an SCV allowing for further replication, and the migration of infected phagocytes allows for the spread of the invading bacteria, and subsequent circulation in the host’s bloodstream to several tissues including the spleen, liver, and in extreme cases the brain.
Antibiotic Resistance and Treatment
Risks Associated With S. enterica infections
Risk Factors

Disease Prevention Measures
References
[1]
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2015, Kenyon College.
