Salmonella enterica serovar Typhimurium: Difference between revisions
No edit summary |
No edit summary |
||
| (65 intermediate revisions by 15 users not shown) | |||
| Line 1: | Line 1: | ||
{{Conway}} | |||
{{ | |||
[[File:Salmonella_Typhimurium_pathogenesis.jpeg|400px|thumb|right|Scanning electron microscope image of <i>Salmonella Typhimurium</i>. From: Nbcnews.com [http://media1.s-nbcnews.com/j/MSNBC/Components/Photo/_new/g-hlt-090127-salmonella-typhimurium-5p.grid-4x2.jpg]]] | [[File:Salmonella_Typhimurium_pathogenesis.jpeg|400px|thumb|right|Scanning electron microscope image of <i>Salmonella Typhimurium</i>. From: Nbcnews.com [http://media1.s-nbcnews.com/j/MSNBC/Components/Photo/_new/g-hlt-090127-salmonella-typhimurium-5p.grid-4x2.jpg]]] | ||
| Line 8: | Line 6: | ||
| Domain = [[Bacteria]] | | Domain = [[Bacteria]] | ||
| Phylum = [[Proteobacteria]] | | Phylum = [[Proteobacteria]] | ||
| Class = | | Class = Gammaproteobacteria | ||
| Order = [[Enterobacteriales]] | | Order = [[Enterobacteriales]] | ||
| Family = | | Family = Enterobacteriaceae | ||
| Genus = [[Salmonella]] | | Genus = [[Salmonella]] | ||
| Species = [[enterica]] | | Species = [[S. enterica]] | ||
| serotype = [[Typhimurium]] | | serotype = [[Typhimurium]] | ||
<br> | <br> | ||
{|| height="10" bgcolor="#FFDF95" |'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=90371&lvl=3&p=mapview&p=has_linkout&p=blast_url&p=genome_blast&lin=f&keep=1&srchmode=1&unlock Taxonomy] Genome: <font size="2">[http://www.ncbi.nlm.nih.gov/genome/?term=salmonella+enterica Genome]</font>'''|} | {|| height="10" bgcolor="#FFDF95" |'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=90371&lvl=3&p=mapview&p=has_linkout&p=blast_url&p=genome_blast&lin=f&keep=1&srchmode=1&unlock Taxonomy] Genome: <font size="2">[http://www.ncbi.nlm.nih.gov/genome/?term=salmonella+enterica Genome]</font>'''|} | ||
===Description=== | ===Description=== | ||
Being zoonotic, <i>Salmonella enterica</i> subsp. enterica serovar Typhimurium ST313 is a rod-shaped, Gram-negative, flagellated facultative anaerobe that is mostly present in the mammalian GI tract [[#References|[1]]]. It is a Non-Typhoidal <i>Salmonella</i> serotype that causes diarrhea and paediatric blood stream infections (bacteremia). It is one of the few emerging invasive strains of Non-Typhoidal <i>Salmonella</i> (iNTS)[[#References|[2]]]. This specific strain, Typhimurium ST313, was found prevalent in the sub-Saharan African population, especially among those with high prevalence of HIV, with an associated case fatality of 20–25% [[#References|[1]]]. In relation to diarrhoeal disease, non-typhoidal <i>Salmonella</i> can exploit the gut mucosal inflammatory response that accompanies infection in immunocompetent individuals to gain a selective advantage over the resident gut microbiota in the inflamed gut lumen. The clinical features of this iNTS disease in Africa are diverse; the disease differs from other strains of <i>Salmonella</i>. Fever, hepatosplenomegaly, and respiratory symptoms are common. Features of enterocolitis are often absent [[#References|[1]]]. Treatment consists of the administration of antibiotics; however, with caution of possible antimicrobial resistance. Whole-genome sequence analysis of 129 ST313 strains, isolated during 1988–2010 from 7 countries of sub-Saharan Africa, identified 2 dominant genetic lineages, I and II [[#References|[3]]]. Further studies are still needed to fully understand and control this iNTS strain. | |||
==Pathogenesis== | ==Pathogenesis== | ||
===Transmission/Reservoirs=== | ===Transmission/Reservoirs=== | ||
<i>Salmonella | <i>Salmonella</i> Typhimurium is generally thought to have a wide range of animal hosts, including birds, cattle and many other domesticated animals. However, recent research has led many to believe that the individual variants have a much more narrow range of possible hosts [[#References|[1]]] . The invasive African variant has only been found to infect humans; specifically people with compromised immune systems. Since the beginning of the HIV epidemic, through the Sub-Saharan, invasive African <i>Salmonella</i> Typhimurium has been discovered among the most severe HIV victims as a human-to-human pathogen [[#References|[2]]]. | ||
<br><br> | <br><br> | ||
Common modes of infection for <i>Salmonella</i> Typhimurium are by food-borne transmission including processed foods such as chocolate and jalapeño peppers [[#References|[1]]]. These are especially common in hospital settings [[#References|[3]]]. Many of the most severe infections are believed to be hospital-acquired. Patients with severe onset HIV are at the highest risk of infection while malnourished children are the next most susceptible group [[#References|[3]]]. | |||
<br><br> | <br><br> | ||
Along with susceptible groups stated above, other links have been found between those suffering from malaria, sickle cell anemia, and people recently treated with gastric acid suppression and the acquisition of <i>Salmonella</i> Typhimurium. | |||
===Incubation/Colonization=== | ===Incubation/Colonization=== | ||
Invasive, African <i>Salmonella | Invasive, African <i>Salmonella</i> Typhimurium does not typical cause disease in healthy individuals, but those that are immunocompromised due to other diseases (eg HIV) or just for lack of overall nutrition, are much more likely to be infected by the pathogen [[#References|[4]]]. There is no conclusive data on whether the invasive or gastrointestinal forms of <i>Salmonella</i> Typhimurium are actually two different pathogens, or if their modes of transition, colonization, or incubation differ [[#References|[1]]]. | ||
===Epidemiology=== | ===Epidemiology=== | ||
Incidents of <i>Salmonella</i> Typhimurium occur largely in Sub-Saharan Africa, with the highest incident rates occurring during or just after the specific area's rainy season. The median ages of those infected are 32 for adults and 22 months for children [[#References|[5]]]. Overall, there has not been an in-depth study done into the epidemiology of either invasive or non-invasive non-typhoidal <i>Salmonella</i> strains [[#References|[1]]]. | |||
===Virulence Factors=== | ===Virulence Factors=== | ||
The most alarming virulence factor of invasive <i>Salmonella | In <i>Salmonella</i>, several genes have been identified to encode various virulence factors. In the bacterial genome, these genes are located relatively closely to one another in groups known as <i>Salmonella</i> Pathogenicity Islands (SPIs). Currently, there are about 21 identified SPIs (from SPI-1 to SPI-21) for <i>Salmonella</i>. For <i>Salmonella</i> Typhimurium specifically, there are 12 known SPIs presenting different virulence factors: SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-6, SPI-9, SPI-11, SPI-12, SPI-13, SPI-14, and SPI-16 [[#References|[9]]]. | ||
The most alarming virulence factor of invasive <i>Salmonella</i> Typhimurium is the rapid acquisition of drug-resistance, which has led to multidrug-resistant “super” strains unaffected by common antibiotics such as ampicillin, cotrimoxazole, and chloramphenicol [[#References|[4]]]. Invasive <i>Salmonella</i> Typhimurium also has many similar virulence factors to <i>Salmonella</i> Typhi. These include type III secretion systems, Vi antigen, many surface polysaccharides, and flagella [[#References|[5]]]. | |||
==Clinical Features== | ==Clinical Features== | ||
Acute gastroenteritis is the most common symptom in infected patients. This causes diarrhea, abdominal cramping, fever, and vomiting. Fever will usually subside in 72 hours, with bloody diarrhea lasting between three and seven days. These effects can be more severe or prolonged in children and the elderly. Bacteremia, or the spread of the pathogen into the blood stream, generally occurs in 5-10% of cases and can lead to more severe symptoms such as meningitis and infections of the bones and joints. This can be especially dangerous in immunocompromised patients such as those suffering from HIV or | Acute gastroenteritis is the most common symptom in infected patients. This causes diarrhea, abdominal cramping, fever, and vomiting. Fever will usually subside in 72 hours, with bloody diarrhea lasting between three and seven days. These effects can be more severe or prolonged in children and the elderly. Bacteremia, or the spread of the pathogen into the blood stream, generally occurs in 5-10% of cases and can lead to more severe symptoms such as meningitis and infections of the bones and joints. This can be especially dangerous in immunocompromised patients such as those suffering from HIV or malaria [[#References|[6]]]. | ||
==Diagnosis== | ==Diagnosis== | ||
The classical method for confirming a | The classical method for confirming a <i>Salmonella</i> infection was by testing for the presence of antibodies using a Widal aggulation test. This method is highly unreliable and in an experiment only tested positive on 38.5% of confirmed <i>Salmonella</i> infections [[#References|[7]]]. Since then, a new method for detecting antibodies has been developed using an enzyme immunoassay for IgM, IgG, and IgA antibodies. This method uses a commercially available lipopolysaccharaide for <i>S.</i> Typhimurium and <i>S.</i> enteritidis. This method produced a much better 88.5% positive rate of confirmed cases. An alternative culturing method can be used to confirm diagnosis by tablet diffusion on Danish Blood Agar with Rosco Neosensitabs. If this test identifies a nalidixic acid resistance, an E-test can also be used to confirm diagnosis [[#References|[8]]]. | ||
==Treatment== | ==Treatment== | ||
From 1998 through 2002, treatment for adults with suspected sepsis was empirical antibiotic treatment that included chloramphenicol and benzylpenicillin. In 2002, chloramphenicol was replaced with oral ciprofloxacin. For adults who did not respond to initial treatments or were unable to take oral medication, they were prescribed parenteral gentamicin. In 1998 to 2002, empirical treatment for childhood sepsis consists of chloramphenicol. Parenteral gentamicin was also added to the treatment from 2002 onwards [[#References|[4]]]. Other antibiotics used for treatment are penicillin with chloramphenicol or ampicillin with gentamicin [[#References|[3]]]. | |||
For multidrug-resistant strains of invasive non-typhoidal <i>Salmonella</i>, treatment drugs include third generation cephalosporins and fluoroquinolone (i.e. ciprofloxacin) [[#References|[1]]]. | |||
In the context of HIV, recurrent invasive non-typhoidal <i>Salmonella</i> disease declined after being treated with antiretroviral therapy [[#References|[1]]]. | |||
==Prevention== | ==Prevention== | ||
===Risk Factors=== | |||
Environmental risk factors for both adults and children include rainy seasons when drinking water sources have the highest concentrations of fecal organisms. Animal contact is a risk factor especially for children. Hospitals are also places where patients can contract hospital-acquired infections [[#References|[3]]]. | |||
Host risk factors include age, exposure to antimicrobial agents, malaria and anemia, malnutrition, HIV infection, gastric acid suppression, sickle cell disease, and schistosomiasis. Children and infants under the age of 3 years are particularly at risk for invasive iNTS disease. Adults above 50 years old in industrialized countries are more susceptible to endovascular infections with iNTS, and younger adults are dominated by HIV-associated iNTS disease. Malnutrition and other previous/current diseases increase the risk of iNTS disease by reducing immune system responses and leaving the body vulnerable to infection [[#References|[3]]]. | |||
Preventive measures include vaccines, improved hygiene and sanitation (washing hands), improved nutrition (safe drinking water and proper food preparation and storage), malaria control, and anti-retroviral therapy programs [[#References|[1]]]. | |||
==Host Immune Response== | |||
===Gastric Innate Immune Response=== | |||
First, a thick mucus layer provides a barrier against <i>Salmonella Typhimurium</i>. Cells of the GI tract also secrete “antibacterial-like” antimicrobial peptides that disrupts the integrity and composition of the bacteria’s cell walls. Yet, once the pathogen breaks through this physical barrier, it starts to invade M cells or enterocytes. This triggers the next line of defense to appear: macrophages and dendritic cells phagocytosing, killing, and signaling other immune cells to the site of infection. Yet, the pathogen prefers to be engulfed in order to continue its infection. The internal survival of the pathogen eventually leads to mucosal inflammation, neutrophil influxes, and more inflammation. However, <i>Salmonella Typhimurium</i> uses the inflammation that it caused to its advantage by weakening the human gut microbiota and dictate the infections there [[#References|[11]]]. | |||
===Entry/Localization Dependent=== | |||
Where and how <i>Salmonella Typhimurium</i> triggers an immune response still remains controversial. One entry way involves invading microfold cells (M cells) with the help of its high flagellated motility. M cell invasion is just the first step of <i>Salmonella Typhimurium</i> infection because at later times, the pathogen is able to move in the mucous layer and to reach and invade enterocytes. | |||
The other route involves the phagocytosis from dendritic cells, who follows chemokines until the find Toll-like receptors (TLR). This contributes to the wide distribution of <i>Salmonella Typhimurium</i>, helping it to be classified as a systemic pathogen [[#References|[10]]]. | |||
<i>Salmonella Typhimurium</i> invades host cells by injecting virulence factors into target cells and driving cytoskeleton rearrangements and phagocytosis via the expression of a type-three secretion system (TTSS) encoded within the Salmonella pathogenicity island I (SPI-1). | |||
The type of host immune response induced by <i>Salmonella Typhimurium</i> depends on the location of infection. Depending on the route of Salmonella entry, systemic or mucosal responses are initiated. <i>Salmonella</i> entry via M cells delivers bacteria directly into the subepithelial dome of Peyer’s patches (PPs). PPs are absolutely required for the initiation of a specific IgA response to <i>Salmonella</i>, whereas they are dispensable for a systemic response [[#References|[10]]]. | |||
This specific IgA response to <i>Salmonella</i> is a result of the immune system sensing the presence of bacteria that invade M cells and preparing against their massive entrance by secreting specific SIgAs, which has an important function in anti-<i>Salmonella</i> immunity. Intestinal IgAs play a major role in protecting mucosal surfaces against colonization and invasion by pathogenic microorganisms but also in limiting commensal bacteria to the intestinal lumen. Secretory IgAs represent a first line of defense against mucosal pathogens by limiting the entrance of bacteria, a process named “immune exclusion.” This compartmentalization of the immune response could allow the fast and specific generation of anti-<i>Salmonella</i> IgAs that are required to protect the host against subsequent challenges with the pathogen [[#References|[10]]]. | |||
At the same time capture of <i>Salmonella</i> by intraepithelial DCs can deliver the <i>Salmonella</i> to the LP and the mesenteric lymph nodes (MLNs). This bacteria’s distribution directed the type of induced immune response, the production of SIgAs, and subsequent protective immunity [[#References|[10]]]. | |||
== Damage Response Framework== | |||
[[File:Salmonella.png|left|thumb|alt=Example alt text|Classification Salmonella Thyphimurium according to the Damage-Response Framework. | |||
Invasive African Salmonella Thyphimurium has only been known to infect humans, specifically HIV-infected humans with weak immune responses. As with other class 3 pathogens, Salmonella Thyphimurium causes damage at both ends of the continuum of immune responses. The pathogen promotes its own phagocytosis as the host initiates an inflammatory response, causing damage as the host’s immune response strengthens. However, the damage will not be as severe as that resulting from a weak host response, as this pathogen usually infects immunocompromised individuals.]] | |||
''Salmonella enterica serovar Typhimurium'' can be classified as a '''Class 3 pathogen,''' based on the Damage Response Framework classification system, because it causes a response in all host cells along the continuum of host immune response, but causes significantly more damage in the setting of weak or strong host immune responses. | |||
It poses the most significant threat to immunocompromised hosts because it faces little colonization resistance from the host immune system. This particular strain of ''S. typhurium'' has been found to cause serious infection in HIV-positive individuals, and its risk factors include malnutrition, sickle cell disease, and other immune system-hindering factors. | |||
Additionally, it causes serious damage in hosts with strong immune responses because the host’s innate immune response includes phagocytosis of the pathogen by macrophages and stimulation of inflammation, both of which aid the colonization of ''S. typhimurium throughout the body.'' | |||
==References== | ==References== | ||
1. Chen HM, Wang Y, Su LH, Chiu CH: Nontyphoid Salmonella Infection: Microbiology, Clinical Features, and Antimicrobial Therapy. <i>Pediatrics & Neonatology</i> 2013, 54:3:147-152.<br> | 1. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA: Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. <i>Lancet</i> 2012. 379:9835::2489-99. | ||
2. Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, MacLennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G: Intracontinental spread of human invasive <i>Salmonella</i> Typhimurium pathovariants in sub-Saharan Africa. <i>Nature Genetics</i> 2012. 44:1215–1221. | |||
3. Morpeth SC, Ramadhani HO, Crump JA. Invasive Non-Typhi Salmonella Disease in Africa. <i>Clin Infect Dis.</i> (2009) 49:4:606-611. | |||
4. Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, Zijlstra EE, Heyderman RS, Hart CA, Molyneux ME: Epidemics of Invasive <i>Salmonella enterica</i> serovar Enteritidis and <i>S. enterica</i> serovar Typhimurium Infection Associated with Multidrug Resistance among Adults and Children in Malawi. <i>Clin Infect Dis.</i> 2008. 46:7:963-969. | |||
5. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ: Host–Pathogen Interaction in Invasive Salmonellosis. PLoS Pathog 2012. 8:10. | |||
6. Chen HM, Wang Y, Su LH, Chiu CH: Nontyphoid Salmonella Infection: Microbiology, Clinical Features, and Antimicrobial Therapy. <i>Pediatrics & Neonatology</i> 2013, 54:3:147-152.<br> | |||
7. Isomaki O, Vuento R, Granfors K: Serological Diagnosis of Salmonella Infections by Enzyme Immunoassay. <i>Lancet</i> 1989, 333:8652:1411-1414.<br> | |||
8. Helms M, Vastrup P, Gerner-Smidt P, Mølbak K: Excess Mortality Associated with Antimicrobial Drug-Resistant <i>Salmonella</i> Typhimurium. <i>Emerging Infectious Diseases</i> 2002, 8:5:490-495.<br> | |||
9. López FE, Pescaretti MM, Morero R, Delgado MA: <i>Salmonella</i> Typhimurium general virulence factors: A battle of David against Goliath?. <i> Food Research International </i> 2011, 45: 842-851. | |||
10. Chiavelli A, Martinoli C, Rescigno M: Entry Route of Salmonella typhimurium Directs the Type of Induced Immune Response. <i>Immunity</i> 2007. 27: 975–984. | |||
11. Broz P, Monack DM, Ohlson MB: Innate immune response to Salmonella typhimurium, a model enteric pathogen. <i>Gut Microbes</i> 2012. 3(2): 62-70. | |||
12. Casadevall, A., & Pirofski, L. (n.d.). The damage-response framework of microbial pathogenesis. <i>Nat Rev Micro Nature Reviews Microbiology</i> 2003. 17-24. | |||
Created by Nathan Sethman, Robertson Bootes Beasley, Amy Vu, Teresa Vu, Danielle Lewis, Elizabeth Fish, and Erika Cummings, students of Tyrrell Conway at the University of Oklahoma.<br> | |||
Latest revision as of 16:41, 11 February 2016


Etiology/Bacteriology
Taxonomy
| Domain = Bacteria
| Phylum = Proteobacteria
| Class = Gammaproteobacteria
| Order = Enterobacteriales
| Family = Enterobacteriaceae
| Genus = Salmonella
| Species = S. enterica
| serotype = Typhimurium
Description
Being zoonotic, Salmonella enterica subsp. enterica serovar Typhimurium ST313 is a rod-shaped, Gram-negative, flagellated facultative anaerobe that is mostly present in the mammalian GI tract [1]. It is a Non-Typhoidal Salmonella serotype that causes diarrhea and paediatric blood stream infections (bacteremia). It is one of the few emerging invasive strains of Non-Typhoidal Salmonella (iNTS)[2]. This specific strain, Typhimurium ST313, was found prevalent in the sub-Saharan African population, especially among those with high prevalence of HIV, with an associated case fatality of 20–25% [1]. In relation to diarrhoeal disease, non-typhoidal Salmonella can exploit the gut mucosal inflammatory response that accompanies infection in immunocompetent individuals to gain a selective advantage over the resident gut microbiota in the inflamed gut lumen. The clinical features of this iNTS disease in Africa are diverse; the disease differs from other strains of Salmonella. Fever, hepatosplenomegaly, and respiratory symptoms are common. Features of enterocolitis are often absent [1]. Treatment consists of the administration of antibiotics; however, with caution of possible antimicrobial resistance. Whole-genome sequence analysis of 129 ST313 strains, isolated during 1988–2010 from 7 countries of sub-Saharan Africa, identified 2 dominant genetic lineages, I and II [3]. Further studies are still needed to fully understand and control this iNTS strain.
Pathogenesis
Transmission/Reservoirs
Salmonella Typhimurium is generally thought to have a wide range of animal hosts, including birds, cattle and many other domesticated animals. However, recent research has led many to believe that the individual variants have a much more narrow range of possible hosts [1] . The invasive African variant has only been found to infect humans; specifically people with compromised immune systems. Since the beginning of the HIV epidemic, through the Sub-Saharan, invasive African Salmonella Typhimurium has been discovered among the most severe HIV victims as a human-to-human pathogen [2].
Common modes of infection for Salmonella Typhimurium are by food-borne transmission including processed foods such as chocolate and jalapeño peppers [1]. These are especially common in hospital settings [3]. Many of the most severe infections are believed to be hospital-acquired. Patients with severe onset HIV are at the highest risk of infection while malnourished children are the next most susceptible group [3].
Along with susceptible groups stated above, other links have been found between those suffering from malaria, sickle cell anemia, and people recently treated with gastric acid suppression and the acquisition of Salmonella Typhimurium.
Incubation/Colonization
Invasive, African Salmonella Typhimurium does not typical cause disease in healthy individuals, but those that are immunocompromised due to other diseases (eg HIV) or just for lack of overall nutrition, are much more likely to be infected by the pathogen [4]. There is no conclusive data on whether the invasive or gastrointestinal forms of Salmonella Typhimurium are actually two different pathogens, or if their modes of transition, colonization, or incubation differ [1].
Epidemiology
Incidents of Salmonella Typhimurium occur largely in Sub-Saharan Africa, with the highest incident rates occurring during or just after the specific area's rainy season. The median ages of those infected are 32 for adults and 22 months for children [5]. Overall, there has not been an in-depth study done into the epidemiology of either invasive or non-invasive non-typhoidal Salmonella strains [1].
Virulence Factors
In Salmonella, several genes have been identified to encode various virulence factors. In the bacterial genome, these genes are located relatively closely to one another in groups known as Salmonella Pathogenicity Islands (SPIs). Currently, there are about 21 identified SPIs (from SPI-1 to SPI-21) for Salmonella. For Salmonella Typhimurium specifically, there are 12 known SPIs presenting different virulence factors: SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-6, SPI-9, SPI-11, SPI-12, SPI-13, SPI-14, and SPI-16 [9].
The most alarming virulence factor of invasive Salmonella Typhimurium is the rapid acquisition of drug-resistance, which has led to multidrug-resistant “super” strains unaffected by common antibiotics such as ampicillin, cotrimoxazole, and chloramphenicol [4]. Invasive Salmonella Typhimurium also has many similar virulence factors to Salmonella Typhi. These include type III secretion systems, Vi antigen, many surface polysaccharides, and flagella [5].
Clinical Features
Acute gastroenteritis is the most common symptom in infected patients. This causes diarrhea, abdominal cramping, fever, and vomiting. Fever will usually subside in 72 hours, with bloody diarrhea lasting between three and seven days. These effects can be more severe or prolonged in children and the elderly. Bacteremia, or the spread of the pathogen into the blood stream, generally occurs in 5-10% of cases and can lead to more severe symptoms such as meningitis and infections of the bones and joints. This can be especially dangerous in immunocompromised patients such as those suffering from HIV or malaria [6].
Diagnosis
The classical method for confirming a Salmonella infection was by testing for the presence of antibodies using a Widal aggulation test. This method is highly unreliable and in an experiment only tested positive on 38.5% of confirmed Salmonella infections [7]. Since then, a new method for detecting antibodies has been developed using an enzyme immunoassay for IgM, IgG, and IgA antibodies. This method uses a commercially available lipopolysaccharaide for S. Typhimurium and S. enteritidis. This method produced a much better 88.5% positive rate of confirmed cases. An alternative culturing method can be used to confirm diagnosis by tablet diffusion on Danish Blood Agar with Rosco Neosensitabs. If this test identifies a nalidixic acid resistance, an E-test can also be used to confirm diagnosis [8].
Treatment
From 1998 through 2002, treatment for adults with suspected sepsis was empirical antibiotic treatment that included chloramphenicol and benzylpenicillin. In 2002, chloramphenicol was replaced with oral ciprofloxacin. For adults who did not respond to initial treatments or were unable to take oral medication, they were prescribed parenteral gentamicin. In 1998 to 2002, empirical treatment for childhood sepsis consists of chloramphenicol. Parenteral gentamicin was also added to the treatment from 2002 onwards [4]. Other antibiotics used for treatment are penicillin with chloramphenicol or ampicillin with gentamicin [3].
For multidrug-resistant strains of invasive non-typhoidal Salmonella, treatment drugs include third generation cephalosporins and fluoroquinolone (i.e. ciprofloxacin) [1].
In the context of HIV, recurrent invasive non-typhoidal Salmonella disease declined after being treated with antiretroviral therapy [1].
Prevention
Risk Factors
Environmental risk factors for both adults and children include rainy seasons when drinking water sources have the highest concentrations of fecal organisms. Animal contact is a risk factor especially for children. Hospitals are also places where patients can contract hospital-acquired infections [3].
Host risk factors include age, exposure to antimicrobial agents, malaria and anemia, malnutrition, HIV infection, gastric acid suppression, sickle cell disease, and schistosomiasis. Children and infants under the age of 3 years are particularly at risk for invasive iNTS disease. Adults above 50 years old in industrialized countries are more susceptible to endovascular infections with iNTS, and younger adults are dominated by HIV-associated iNTS disease. Malnutrition and other previous/current diseases increase the risk of iNTS disease by reducing immune system responses and leaving the body vulnerable to infection [3].
Preventive measures include vaccines, improved hygiene and sanitation (washing hands), improved nutrition (safe drinking water and proper food preparation and storage), malaria control, and anti-retroviral therapy programs [1].
Host Immune Response
Gastric Innate Immune Response
First, a thick mucus layer provides a barrier against Salmonella Typhimurium. Cells of the GI tract also secrete “antibacterial-like” antimicrobial peptides that disrupts the integrity and composition of the bacteria’s cell walls. Yet, once the pathogen breaks through this physical barrier, it starts to invade M cells or enterocytes. This triggers the next line of defense to appear: macrophages and dendritic cells phagocytosing, killing, and signaling other immune cells to the site of infection. Yet, the pathogen prefers to be engulfed in order to continue its infection. The internal survival of the pathogen eventually leads to mucosal inflammation, neutrophil influxes, and more inflammation. However, Salmonella Typhimurium uses the inflammation that it caused to its advantage by weakening the human gut microbiota and dictate the infections there [11].
Entry/Localization Dependent
Where and how Salmonella Typhimurium triggers an immune response still remains controversial. One entry way involves invading microfold cells (M cells) with the help of its high flagellated motility. M cell invasion is just the first step of Salmonella Typhimurium infection because at later times, the pathogen is able to move in the mucous layer and to reach and invade enterocytes. The other route involves the phagocytosis from dendritic cells, who follows chemokines until the find Toll-like receptors (TLR). This contributes to the wide distribution of Salmonella Typhimurium, helping it to be classified as a systemic pathogen [10].
Salmonella Typhimurium invades host cells by injecting virulence factors into target cells and driving cytoskeleton rearrangements and phagocytosis via the expression of a type-three secretion system (TTSS) encoded within the Salmonella pathogenicity island I (SPI-1). The type of host immune response induced by Salmonella Typhimurium depends on the location of infection. Depending on the route of Salmonella entry, systemic or mucosal responses are initiated. Salmonella entry via M cells delivers bacteria directly into the subepithelial dome of Peyer’s patches (PPs). PPs are absolutely required for the initiation of a specific IgA response to Salmonella, whereas they are dispensable for a systemic response [10].
This specific IgA response to Salmonella is a result of the immune system sensing the presence of bacteria that invade M cells and preparing against their massive entrance by secreting specific SIgAs, which has an important function in anti-Salmonella immunity. Intestinal IgAs play a major role in protecting mucosal surfaces against colonization and invasion by pathogenic microorganisms but also in limiting commensal bacteria to the intestinal lumen. Secretory IgAs represent a first line of defense against mucosal pathogens by limiting the entrance of bacteria, a process named “immune exclusion.” This compartmentalization of the immune response could allow the fast and specific generation of anti-Salmonella IgAs that are required to protect the host against subsequent challenges with the pathogen [10].
At the same time capture of Salmonella by intraepithelial DCs can deliver the Salmonella to the LP and the mesenteric lymph nodes (MLNs). This bacteria’s distribution directed the type of induced immune response, the production of SIgAs, and subsequent protective immunity [10].
Damage Response Framework
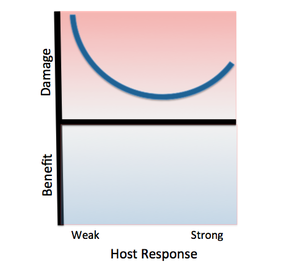
Salmonella enterica serovar Typhimurium can be classified as a Class 3 pathogen, based on the Damage Response Framework classification system, because it causes a response in all host cells along the continuum of host immune response, but causes significantly more damage in the setting of weak or strong host immune responses.
It poses the most significant threat to immunocompromised hosts because it faces little colonization resistance from the host immune system. This particular strain of S. typhurium has been found to cause serious infection in HIV-positive individuals, and its risk factors include malnutrition, sickle cell disease, and other immune system-hindering factors.
Additionally, it causes serious damage in hosts with strong immune responses because the host’s innate immune response includes phagocytosis of the pathogen by macrophages and stimulation of inflammation, both of which aid the colonization of S. typhimurium throughout the body.
References
1. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA: Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 2012. 379:9835::2489-99.
2. Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, MacLennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G: Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nature Genetics 2012. 44:1215–1221.
3. Morpeth SC, Ramadhani HO, Crump JA. Invasive Non-Typhi Salmonella Disease in Africa. Clin Infect Dis. (2009) 49:4:606-611.
4. Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, Zijlstra EE, Heyderman RS, Hart CA, Molyneux ME: Epidemics of Invasive Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium Infection Associated with Multidrug Resistance among Adults and Children in Malawi. Clin Infect Dis. 2008. 46:7:963-969.
5. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ: Host–Pathogen Interaction in Invasive Salmonellosis. PLoS Pathog 2012. 8:10.
6. Chen HM, Wang Y, Su LH, Chiu CH: Nontyphoid Salmonella Infection: Microbiology, Clinical Features, and Antimicrobial Therapy. Pediatrics & Neonatology 2013, 54:3:147-152.
7. Isomaki O, Vuento R, Granfors K: Serological Diagnosis of Salmonella Infections by Enzyme Immunoassay. Lancet 1989, 333:8652:1411-1414.
8. Helms M, Vastrup P, Gerner-Smidt P, Mølbak K: Excess Mortality Associated with Antimicrobial Drug-Resistant Salmonella Typhimurium. Emerging Infectious Diseases 2002, 8:5:490-495.
9. López FE, Pescaretti MM, Morero R, Delgado MA: Salmonella Typhimurium general virulence factors: A battle of David against Goliath?. Food Research International 2011, 45: 842-851.
10. Chiavelli A, Martinoli C, Rescigno M: Entry Route of Salmonella typhimurium Directs the Type of Induced Immune Response. Immunity 2007. 27: 975–984.
11. Broz P, Monack DM, Ohlson MB: Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 2012. 3(2): 62-70.
12. Casadevall, A., & Pirofski, L. (n.d.). The damage-response framework of microbial pathogenesis. Nat Rev Micro Nature Reviews Microbiology 2003. 17-24.
Created by Nathan Sethman, Robertson Bootes Beasley, Amy Vu, Teresa Vu, Danielle Lewis, Elizabeth Fish, and Erika Cummings, students of Tyrrell Conway at the University of Oklahoma.
