Secondary Metabolites in the Aspergillus Genus
Introduction
Aspergillus, a genus comprised of filamentous fungi, is a widely studied group of sac fungi containing a diverse array of both beneficial and pathogenic species. Like all fungi, Aspergillus species are eukaryotic and have a cell wall made of chitin. They are heterotrophic; most commonly found in soil habitats, but are also found in marine environments. Some common species include Aspergillus fumigatus, responsible for the highest number of human deaths from fungi, Aspergillus flavus, a destructive agricultural pest, and Aspergillus nidulans, an important model organism for (Gibbons and Rokas 2013). A wide variation in metabolism, reproductive strategies, and survival mechanisms are found among Aspergillus members, rendering them not only as resilient and diverse microbes, but also as important organisms for biological study.
All Aspergillus produce asci (sexual spore producing cells) within ascocarps (circular fruiting bodies). Some members have the ability to reproduce both sexually and asexually, depending on favorable or unfavorable environmental conditions. This reproductive strategy adds to their resilience and commonality in a diverse range of ecosystems. Many Aspergillus species also possess the ability to form symbiotic relationships with other fungi, bacteria, algae, plants, and even insects. That said, many species are also pathogenic, and many species account for a majority of plant and human pathogens.
A defining characteristic of Aspergillus is their ability to produce secondary metabolites in response to environmental parameters, allowing them to adapt to complex and changing environments. These secondary metabolites allow fungi to either increase their own fitness or decrease a surrounding organism’s fitness, ensuring survival and reproduction. Despite significant scholarship surrounding the Aspergillus genus, their production of secondary metabolites remains a relatively unexplored area of study.
The metabolic and reproductive characteristics of Aspergillus, as outlined above, render this genus important both in microbial research and in other biological topics, such as genetics, bioengineering, ecology, and biochemistry.
Aspergillus Metabolism
Primary metabolism of Aspergillus relies on a distinct set of fatty acid synthase complexes that build fatty acid chains from the breakdown of carboxylic acids. During fungal growth and development, primary metabolism generates long fatty acid chains, which are used for cell membrane production and the formation of vesical storage bodies. In contrast, secondary metabolism largely involves polyketide synthases that generate polyketides molecules from distinct acidic compounds. For example, hexanoyl coA is used as a building block for the secondary metabolite sterigmatocystin which has both immunosuppressant abilities and carcinogenic qualities. Distinct synthase complexes used for the biosynthesis of different metabolic compounds ensure complex and precise regulation of both metabolisms, often, it is the case that the activation of secondary metabolism is associated with the inhibition of primary metabolic pathways (Brown et. al. 1996).
Secondary Metabolites
The ability for filamentous fungi to produce secondary metabolites from altered or de novo biosynthetic pathways remains a topic of scientific interest. Both the degree of secondary metabolism use and the diversity of secondary metabolites found in Aspergillus still largely remain unknown. This is largely a result of secondary biosynthetic pathways that remain silent or inactive under normal laboratory conditions (Nutzmann et. al. 2011). Therefore, it is difficult to study the mechanisms and signaling pathways behind secondary metabolism activation. Nevertheless, metabolites produced are molecules essential to pharmaceutical, industrial, and agricultural interests and applications.
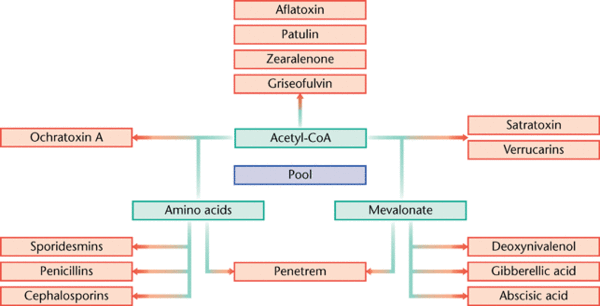
Secondary metabolites are structurally diverse low molecular mass molecules that are not essential for the growth and survival of the producing organism. Differing from the essential role of primary metabolites, secondary metabolites instead serve to increase the fitness of the producing organism or to decrease the fitness of surrounding organisms (Brakhage 2013). The major groups of secondary metabolites found in Aspergillus include polyketides (PKs), ribosomal and nonribosomal peptides (NRPs) and terpenoids (Anderson et. al. 2013). The diverse nature of secondary metabolites is a result of many tailoring enzymes that infer biochemical modifications on an otherwise standard structural polymer (Anderson et. al. 2013). As a result, a complex and intricate level of genetic regulation exists. The genes that code for secondary metabolism are organized in clusters, which encode for primary enzymes, and are flanked by genes that code for regulatory, transporter, or signaling proteins (Brakhage 2013).
A few of the many metabolites synthesized by Aspergillus that have been discovered hold great importance in human application; a few examples include the cholesterol reducing drug lovastin, the antibiotic penicillin, and the pathogenic human toxin aflatoxin (Gibbons and Rokas 2013).
Regulation of Secondary Metabolites
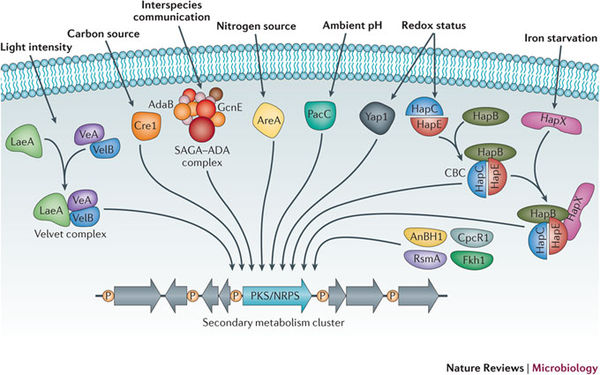
The regulation of secondary metabolism and their subsequent synthesis pathways is controlled by posttranslational modification (Nutzmann et. al. 2011). Studies have shown that histone modification by acetyltransferase complexes of regulatory genes or transcription factors leads to the induction of secondary metabolite production. Similar to DNA methylation, chromatin is reprogrammed which leads to the increased transcription of secondary metabolite gene clusters (Nutzmann et. al. 2011). This complex regulatory network is responsive to various environmental stimuli, including but not limited too, carbon and nitrogen levels, temperature, light, pH, and stimuli from other organisms (Brakhage 2013). These transcription factors are responsive to these stimuli, and when activated, these factors act on the chromatin scaffold comprised of histone proteins, that DNA then wraps around. Phosphorylation, methylation, and acetylation, are all various modifications that are associated with the regulation of secondary metabolism gene clusters (Brakhage 2013). The bacteria, streptomycete, cause an interesting biological example of secondary metabolic induction via mediated histone acetylation. Nutzmann et. al. (2011) outlined the fact that actinomycete bacterial species can induce histone modification in Aspergillus nidulans via the main histone acetytransferase (HAT) complex. It was found that the physical interaction of fungal mycelium and Streptomyces microbes activated the secondary metabolic pathway that resulted in the production of the polyketide orsellinic acid. This metabolic pathway was induced by the activation of the ors gene cluster and the saga/ada complex. Studies such as these illustrate the sensitivity of secondary metabolite production in relation to the surrounding environment, and demonstrate that physical stimuli as well as chemical signals can regulate SM production (Nutzmann et. al. 2011). Likewise, it demonstrates that many complex and relatively unknown ecological factors are involved in the activation of transcription factors inducing secondary metabolite activity.
However diverse the regulation of SM production remains, recent study has shown that a nuclear protein, LaeA is a key master regulatory protein in most Aspergilla species. It has been shown that the knockout of this gene results in widespread reduction of most SM levels. VeA protein has also been identified as a key regulatory protein, increasing expression levels in response to light stimuli. VeA and LaeA interact together to regulate SM production, asexual development, and sporulation. Upregulation of these genes has been found to directly increase secondary metabolite molecules such as sterigmatocystin, terrequinone, and aflatoxins (Fox and Howlett 2008). As demonstrated, the knockout of key genes is an important tool for further elucidation of SM regulation by proteins (transcription factors).
Section 3
Include some current research in each topic, with at least one figure showing data.
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.