Serratia marcescens' different drug resistant efflux pumps: Difference between revisions
(Created page with "{{Uncurated}} =Introduction= "Serratia marcescens" is a gram-negative bacillus bacterium classified as a member of the [http://en.wikipedia.org/wiki/Enterobacteriaceae Entero...") |
No edit summary |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
=Introduction= | =Introduction= | ||
''Serratia marcescens'' is a gram-negative bacillus bacterium classified as a member of the [http://en.wikipedia.org/wiki/Enterobacteriaceae Enterobacteriaceae] family. It is an [http://www.biology-online.org/dictionary/Opportunistic_pathogen opportunistic pathogen] and has been the cause of [http://www.healthline.com/galecontent/nosocomial-infections nosocomial infections] for the past two decades[[#References|[5]]]. ''S. marcescens'' is motile and a facultative anaerobe and is well known for its pink pigmented colonies which are commonly seen on the walls of bathtubs or tiles where soap residue is usually found. Many infections caused by ''S. marcescens'' are difficult to treat due to antibiotic resistance that the bacterium has developed[[#References|[11]]]. Studies have shown that ''S. marcescens'' have multidrug resistance due to the efflux pumps that it maintains[[#References|[3]]]. | |||
[[File:260px-Serratia marcescens.jpg|300px|thumb|center| ''Serratia marcescens''. Retrieved from http://commons.wikimedia.org/wiki/File:Serratia_marcescens.jpg]] | |||
=Efflux Pumps= | |||
[http://en.wikipedia.org/wiki/Efflux_(microbiology) Efflux] pumps in bacterial cells are transport proteins that contribute to the expulsion of toxic substrates from inside the cell to the environment[[#References|[12]]]. Such toxic substrates such as [http://www.drugs.com/erythromycin.html erythromycin], [http://en.wikipedia.org/wiki/Streptomycin streptomycin] and [http://en.wikipedia.org/wiki/Norfloxacin norflaxin], are clinically developed antibiotics that target the cell’s physiological processes. There are five major efflux pumps found in bacterial cells: MF (major facilitator), MATE (multidrug and toxic efflux), RND (resistance-nodulation-division), SMR (small multidrug resistance) and ABC (ATP-binding cassette)[[#References|[12]]]. ''S. marcescens'' is found to have four of the five major efflux pumps. S. marcescens has been characterized to express three RND-type7 and one from the MF, SMR and ABC-type efflux pumps. Efflux pumps play an important role in the drug resistance capabilities of bacteria including ''Escherichia coli'', especially in ''S. marcescens''. | |||
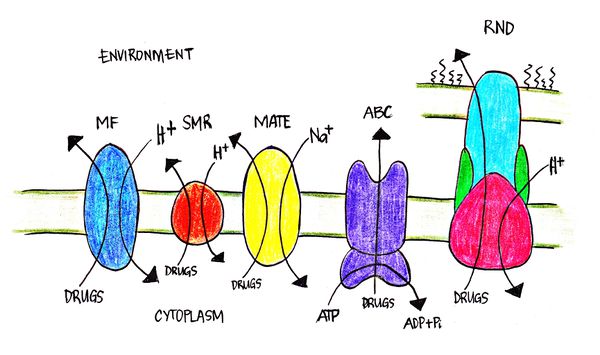
[[File:Efflux pumps.jpg|600px|thumb|center| Figure 2. Different know bacterial efflux pumps in gram-negative bacteria. Shown in the diagram are the flow of substrates (drugs) and energy source for the different efflux pumps.]] | |||
=''Serratia marcescens''' Efflux Pumps= | |||
There are six efflux pumps found in ''S. marcescens'' that have been characterized: | |||
= | ===SdeAB (''Serratia’s'' drug efflux)=== | ||
It is an RND-type (resistance-nodulation-cell division) efflux pump with genes that code for both an MF and RND-type efflux pumps. It uses a proton gradient as an energy source and consists of a tripartite complex protein. It mostly pumps out [http://en.wikipedia.org/wiki/Quinolone fluoroquinolones] and is known to be the major fluoroquinolone pump in ''S. marcescens''.[[#References|[6]]],[[#References|[7]]] | |||
===SdeCDE=== | |||
It is an RND-type efflux pumps that is a combination of one MFP (membrane fusion protein for sdeC) and two RND (sdeDE) pump transporters which uses the proton gradient for energy. This type of efflux pump also exhibits a tripartite complex and is found to have restrictive selectivity only to [http://en.wikipedia.org/wiki/Novobiocin novobiocin]1. It is also found to be the third RND-type pump to be characterized for ''S. marcescens''.[[#References|[1]]] | |||
[ | ===SdeXY=== | ||
It is an RND-type efflux pump which uses a proton gradient as an energy source. It is composed of two components to make up the complex: SdeX, a hydrophilic protein found to be an MFP; SdeY, an integral protein that has many hydrophobic residues and many [http://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] domains. It is resistant to a wide variety of antimicrobial agents and was the first RND-type to be characterized in ''S. marcescens''[[#References|[4]]]. | |||
===SsmE (''Serratia’s'' small multidrug resistance)=== | |||
It is an SMR-type (small multidrug resistance) efflux pumps that is energy-dependent and works as a substrate/H+ [http://en.wikipedia.org/wiki/Antiporter antiporter] and expels substrates out of the cell. It is composed of four transmembrane domains that span the cell membrane. It is particularly resistant to [http://en.wikipedia.org/wiki/Ethidium_bromide ethidium bromide] and [http://en.wikipedia.org/wiki/Acriflavinium_chloride acriflavine] and is known to be an ethidium efflux[[#References|[9]]]. | |||
== | ===SmfY (''Serratia’s'' major facilitator)=== | ||
It is a member of the MF (major facilitator) superfamily transporters that is an energy-dependent efflux pump that contains numerous hydrophobic residues. These residues are long enough to extend around the cell membrane. It is commonly resistant to a known antiseptic called [http://en.wikipedia.org/wiki/Benzalkonium_chloride benzalkonium chloride]. This is the very first MF-type multidrug efflux pump to be discovered[[#References|[10]]]. | |||
===SmdAB (for Serratia MultiDrug resistance)=== | |||
It is an ABC-type (ATP-binding cassette) multidrug efflux pumps which utilizes ATP as an energy source. It is the very first report of a heterodimer that is seen working in a gram-negative bacterium. It is composed of six transmembrane segments and has hydrophilic segments which contain recognized-binding domains. It is found to be resistant to many cytotoxic agents like norflaxin or tetracycline. This was the very first ABC-type multidrug efflux pump to be characterized in ''S. marcescens''[[#References|[8]]]. | |||
'' | =Impact of Multidrug Resistant Efflux Pumps= | ||
The number of patients in recent years has greatly increased due to the drug resistance of ''S. marcescens''[[#References|[3]]]. Life threatening infections such as [http://www.nlm.nih.gov/medlineplus/ency/article/001355.htm septicemia], [http://en.wikipedia.org/wiki/Endocarditis endocarditis] and [http://en.wikipedia.org/wiki/Meningitis meningitis] are only some of the infections caused by ''S. marcescens''[[#References|[11]]]. Due to the evolutionary adaptations of ''S. marcescens'' in drug resistance, such infections stated are extremely difficult to treat. With the characterization and the further studies of the different multidrug efflux pumps, we are able to understand the mechanisms and the distinct antibiotics that ''S. marcescens'' is resistant to which is key in developing inhibitors and other mechanisms to hinder their function[[#References|[11]]]. | |||
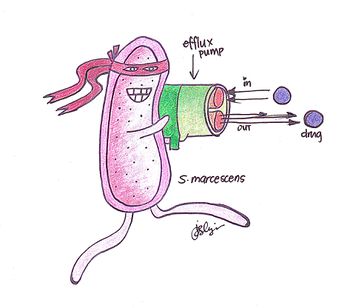
'' | [[File:Serratia efflux pump.jpg|350px|thumb|right| Figure 3. ''S. marcescens'' known to be pretty “bad-ass” towards multiple antibiotics. It regulates its genes to express more efflux pumps due to an increase in antibiotics in the environment.]] | ||
== | =Treatment= | ||
''S. marcescens'' has increased stress in society due to its emerging resistance mechanisms that make its efflux pumps resistant to various antibiotics. Various mechanisms are formulated like controlling the membrane permeability or the production of innovative molecules which are insensitive to these resistant mechanisms for effective treatment[[#References|[2]]]. Recent studies have formulated different strategies in order to elude the drug resistance efflux pumps by doing the following: generating better molecular designs to bypass the efflux activity, directly decreasing the effectiveness of the cell membrane with a channel blocker to induce “traffic jams”, and by blocking the efflux capacity by altering the function of the efflux pump in the bacterial cell wall with inhibition mechanisms[[#References|[2]]]. | |||
=References= | |||
(1)Begic, S., & Worobec, E. (2009). Characterization of the serratia marcescens SdeCDE multidrug efflux pump studied via gene knockout mutagenesis (vol 54, pg 411, 2008).Canadian Journal of Microbiology, 55(9), 1130-1131. doi:10.1139/W09-901 | |||
(2)Mahamoud, A., Chevalier, J., Alibert-Franco, S., Kern, W. V., & Pagès, J. (2007). Antibiotic efflux pumps in gram-negative bacteria: The inhibitor response strategy. Journal of Antimicrobial Chemotherapy, 59(6), 1223-1229. doi:10.1093/jac/dkl493 | |||
( | (3) Chen, J., Lee, E., Kuroda, T., Mizushima, T., & Tsuchiya, T. (2003). Multidrug resistance in serratia marcescens and cloning of genes responsible for the resistance. Biological and Pharmaceutical Bulletin, 26(3), 391-393. doi:10.1248/bpb.26.391 | ||
( | (4) Chen, J., Kuroda, T., Huda, M. N., Mizushima, T., & Tsuchiya, T. (2003). An RND-type multidrug efflux pump SdeXY from serratia marcescens. The Journal of Antimicrobial Chemotherapy, 52(2), 176-179. doi:10.1093/jac/dkg308 | ||
( | (5) HEJAZI, A., & FALKINER, F. R. (1997). Serratia marcescens.Journal of Medical Microbiology, 46(11), 903-912. doi:10.1099/00222615-46-11-903 | ||
( | (6)Kumar, A., & Worobec, E. A.Cloning, sequencing, and characterization of the SdeAB multidrug efflux pump of serratia marcescens American Society for Microbiology. | ||
( | (7) Maseda, H., Hashida, Y., Shirai, A., Omasa, T., & Nakae, T. (2011). Mutation in the sdeS gene promotes expression of the sdeAB efflux pump genes and multidrug resistance in serratia marcescens. Antimicrobial Agents and Chemotherapy, 55(6), 2922-2926. doi:10.1128/AAC.01755-10 | ||
( | (8) Matsuo, T., Chen, J., Minato, Y., Ogawa, W., Mizushima, T., Kuroda, T., & Tsuchiya, T. (2008). SmdAB, a heterodimeric ABC-type multidrug efflux pump, in serratia marcescens.Journal of Bacteriology, 190(2), 648-654. doi:10.1128/JB.01513-07 | ||
( | (9) Minato, Y., Shahcheraghi, F., Ogawa, W., Kuroda, T., & Tsuchiya, T. (2008). Functional gene cloning and characterization of the SsmE multidrug efflux pump from serratia marcescens. Biological & Pharmaceutical Bulletin,31(3), 516-519. doi:10.1248/bpb.31.516 | ||
( | (10) Shahcheraghi, F., Minato, Y., Chen, J., Mizushima, T., Ogawa, W., Kuroda, T., & Tsuchiya, T. (2007). Molecular cloning and characterization of a multidrug efflux pump, SmfY, from serratia marcescens. Biological & Pharmaceutical Bulletin, 30(4), 798-800. | ||
( | (11) Sleigh, J. D. (1983). Antibiotic resistance in serratia marcescens. UK PubMed Central, | ||
( | (12) Webber, M. A., & Piddock, L. J. V. (2003). The importance of efflux pumps in bacterial antibiotic resistance. The Journal of Antimicrobial Chemotherapy, 51(1), 9-11. doi:10.1093/jac/dkg050 | ||
Latest revision as of 23:35, 26 November 2013
Introduction
Serratia marcescens is a gram-negative bacillus bacterium classified as a member of the Enterobacteriaceae family. It is an opportunistic pathogen and has been the cause of nosocomial infections for the past two decades[5]. S. marcescens is motile and a facultative anaerobe and is well known for its pink pigmented colonies which are commonly seen on the walls of bathtubs or tiles where soap residue is usually found. Many infections caused by S. marcescens are difficult to treat due to antibiotic resistance that the bacterium has developed[11]. Studies have shown that S. marcescens have multidrug resistance due to the efflux pumps that it maintains[3].
Efflux Pumps
Efflux pumps in bacterial cells are transport proteins that contribute to the expulsion of toxic substrates from inside the cell to the environment[12]. Such toxic substrates such as erythromycin, streptomycin and norflaxin, are clinically developed antibiotics that target the cell’s physiological processes. There are five major efflux pumps found in bacterial cells: MF (major facilitator), MATE (multidrug and toxic efflux), RND (resistance-nodulation-division), SMR (small multidrug resistance) and ABC (ATP-binding cassette)[12]. S. marcescens is found to have four of the five major efflux pumps. S. marcescens has been characterized to express three RND-type7 and one from the MF, SMR and ABC-type efflux pumps. Efflux pumps play an important role in the drug resistance capabilities of bacteria including Escherichia coli, especially in S. marcescens.
Serratia marcescens' Efflux Pumps
There are six efflux pumps found in S. marcescens that have been characterized:
SdeAB (Serratia’s drug efflux)
It is an RND-type (resistance-nodulation-cell division) efflux pump with genes that code for both an MF and RND-type efflux pumps. It uses a proton gradient as an energy source and consists of a tripartite complex protein. It mostly pumps out fluoroquinolones and is known to be the major fluoroquinolone pump in S. marcescens.[6],[7]
SdeCDE
It is an RND-type efflux pumps that is a combination of one MFP (membrane fusion protein for sdeC) and two RND (sdeDE) pump transporters which uses the proton gradient for energy. This type of efflux pump also exhibits a tripartite complex and is found to have restrictive selectivity only to novobiocin1. It is also found to be the third RND-type pump to be characterized for S. marcescens.[1]
SdeXY
It is an RND-type efflux pump which uses a proton gradient as an energy source. It is composed of two components to make up the complex: SdeX, a hydrophilic protein found to be an MFP; SdeY, an integral protein that has many hydrophobic residues and many transmembrane domains. It is resistant to a wide variety of antimicrobial agents and was the first RND-type to be characterized in S. marcescens[4].
SsmE (Serratia’s small multidrug resistance)
It is an SMR-type (small multidrug resistance) efflux pumps that is energy-dependent and works as a substrate/H+ antiporter and expels substrates out of the cell. It is composed of four transmembrane domains that span the cell membrane. It is particularly resistant to ethidium bromide and acriflavine and is known to be an ethidium efflux[9].
SmfY (Serratia’s major facilitator)
It is a member of the MF (major facilitator) superfamily transporters that is an energy-dependent efflux pump that contains numerous hydrophobic residues. These residues are long enough to extend around the cell membrane. It is commonly resistant to a known antiseptic called benzalkonium chloride. This is the very first MF-type multidrug efflux pump to be discovered[10].
SmdAB (for Serratia MultiDrug resistance)
It is an ABC-type (ATP-binding cassette) multidrug efflux pumps which utilizes ATP as an energy source. It is the very first report of a heterodimer that is seen working in a gram-negative bacterium. It is composed of six transmembrane segments and has hydrophilic segments which contain recognized-binding domains. It is found to be resistant to many cytotoxic agents like norflaxin or tetracycline. This was the very first ABC-type multidrug efflux pump to be characterized in S. marcescens[8].
Impact of Multidrug Resistant Efflux Pumps
The number of patients in recent years has greatly increased due to the drug resistance of S. marcescens[3]. Life threatening infections such as septicemia, endocarditis and meningitis are only some of the infections caused by S. marcescens[11]. Due to the evolutionary adaptations of S. marcescens in drug resistance, such infections stated are extremely difficult to treat. With the characterization and the further studies of the different multidrug efflux pumps, we are able to understand the mechanisms and the distinct antibiotics that S. marcescens is resistant to which is key in developing inhibitors and other mechanisms to hinder their function[11].
Treatment
S. marcescens has increased stress in society due to its emerging resistance mechanisms that make its efflux pumps resistant to various antibiotics. Various mechanisms are formulated like controlling the membrane permeability or the production of innovative molecules which are insensitive to these resistant mechanisms for effective treatment[2]. Recent studies have formulated different strategies in order to elude the drug resistance efflux pumps by doing the following: generating better molecular designs to bypass the efflux activity, directly decreasing the effectiveness of the cell membrane with a channel blocker to induce “traffic jams”, and by blocking the efflux capacity by altering the function of the efflux pump in the bacterial cell wall with inhibition mechanisms[2].
References
(1)Begic, S., & Worobec, E. (2009). Characterization of the serratia marcescens SdeCDE multidrug efflux pump studied via gene knockout mutagenesis (vol 54, pg 411, 2008).Canadian Journal of Microbiology, 55(9), 1130-1131. doi:10.1139/W09-901
(2)Mahamoud, A., Chevalier, J., Alibert-Franco, S., Kern, W. V., & Pagès, J. (2007). Antibiotic efflux pumps in gram-negative bacteria: The inhibitor response strategy. Journal of Antimicrobial Chemotherapy, 59(6), 1223-1229. doi:10.1093/jac/dkl493
(3) Chen, J., Lee, E., Kuroda, T., Mizushima, T., & Tsuchiya, T. (2003). Multidrug resistance in serratia marcescens and cloning of genes responsible for the resistance. Biological and Pharmaceutical Bulletin, 26(3), 391-393. doi:10.1248/bpb.26.391
(4) Chen, J., Kuroda, T., Huda, M. N., Mizushima, T., & Tsuchiya, T. (2003). An RND-type multidrug efflux pump SdeXY from serratia marcescens. The Journal of Antimicrobial Chemotherapy, 52(2), 176-179. doi:10.1093/jac/dkg308
(5) HEJAZI, A., & FALKINER, F. R. (1997). Serratia marcescens.Journal of Medical Microbiology, 46(11), 903-912. doi:10.1099/00222615-46-11-903
(6)Kumar, A., & Worobec, E. A.Cloning, sequencing, and characterization of the SdeAB multidrug efflux pump of serratia marcescens American Society for Microbiology.
(7) Maseda, H., Hashida, Y., Shirai, A., Omasa, T., & Nakae, T. (2011). Mutation in the sdeS gene promotes expression of the sdeAB efflux pump genes and multidrug resistance in serratia marcescens. Antimicrobial Agents and Chemotherapy, 55(6), 2922-2926. doi:10.1128/AAC.01755-10
(8) Matsuo, T., Chen, J., Minato, Y., Ogawa, W., Mizushima, T., Kuroda, T., & Tsuchiya, T. (2008). SmdAB, a heterodimeric ABC-type multidrug efflux pump, in serratia marcescens.Journal of Bacteriology, 190(2), 648-654. doi:10.1128/JB.01513-07
(9) Minato, Y., Shahcheraghi, F., Ogawa, W., Kuroda, T., & Tsuchiya, T. (2008). Functional gene cloning and characterization of the SsmE multidrug efflux pump from serratia marcescens. Biological & Pharmaceutical Bulletin,31(3), 516-519. doi:10.1248/bpb.31.516
(10) Shahcheraghi, F., Minato, Y., Chen, J., Mizushima, T., Ogawa, W., Kuroda, T., & Tsuchiya, T. (2007). Molecular cloning and characterization of a multidrug efflux pump, SmfY, from serratia marcescens. Biological & Pharmaceutical Bulletin, 30(4), 798-800.
(11) Sleigh, J. D. (1983). Antibiotic resistance in serratia marcescens. UK PubMed Central,
(12) Webber, M. A., & Piddock, L. J. V. (2003). The importance of efflux pumps in bacterial antibiotic resistance. The Journal of Antimicrobial Chemotherapy, 51(1), 9-11. doi:10.1093/jac/dkg050