Sphingomonas spp: Agents of Bioremediation and Pathogenesis: Difference between revisions
| Line 32: | Line 32: | ||
==Breakdown of PAHs== | ==Breakdown of PAHs== | ||
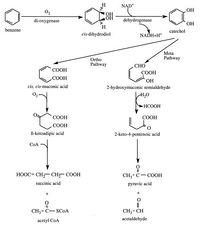
[[File:PAHBreakItDown.jpg|thumb|left||200px|Figure 5. Microbial breakdown of an aromatic ring through <i>ortho</i> or <i>meta</i> cleavage [17].]] | [[File:PAHBreakItDown.jpg|thumb|left||200px|Figure 5. Microbial breakdown of an aromatic ring through <i>ortho</i> or <i>meta</i> cleavage [17].]] | ||
Microbial PAH degradation typically begins with oxidization to <i>cis</i>-dihydrodiols. This ring breakage is energetically costly and is therefore usually the rate-limiting step of the process. This initial oxidation integrates atmospheric oxygen into the substrate (therefore, to utilize this pathway bacteria must oxygen | Microbial PAH degradation typically begins with oxidization to <i>cis</i>-dihydrodiols. This ring breakage is energetically costly and is therefore usually the rate-limiting step of the process. This initial oxidation integrates atmospheric oxygen into the substrate (therefore, to utilize this pathway bacteria must be in contact with oxygen). Bacterial dioxygenase enzymes allow them to break the aromatic nucleus and insert the two oxygen atoms. Once the dihydrodiols are produced they will be re-aromatized using a <i>cis</i>-dihydrodiol dehydrogenase to produce a dihydroxylated intermediate. Continued oxidation of the cis-dihydrodiols eventually results in catechols, which can be targeted by other dioxygenases that cause enzyme-induced cleavage of the aromatic ring. | ||
Two pathways of catechol degradation exist: the <i>ortho</i> pathway cleaves the bond between the two carbons carrying the two hydroxyl groups to produce <i>cis,cis</i>-muconic acid, whereas the <i>meta</i> pathway proceeds through cleavage of the bond between a hydroxyl-bearing carbon and the adjacent carbon with a hydroxyl group (Figure 5). The rest of the intermediates in the pathway are continuously processed into components of biosynthesis and energy. In the course of the reaction, carbon dioxide and water are also released. In this manner, the rest of the rings of a PAH will then be attacked; once one ring is reduced to pyruvic acid and carbon dioxide, the process will move onto the adjacent ring. The more rings are present in a compound, however, the harder it is for the microbe to degrade it due to its aforementioned hydrophobicity [17]. | Two pathways of catechol degradation exist: the <i>ortho</i> pathway cleaves the bond between the two carbons carrying the two hydroxyl groups to produce <i>cis,cis</i>-muconic acid, whereas the <i>meta</i> pathway proceeds through cleavage of the bond between a hydroxyl-bearing carbon and the adjacent carbon with a hydroxyl group (Figure 5). The rest of the intermediates in the pathway are continuously processed into components of biosynthesis and energy. In the course of the reaction, carbon dioxide and water are also released. In this manner, the rest of the rings of a PAH will then be attacked; once one ring is reduced to pyruvic acid and carbon dioxide, the process will move onto the adjacent ring. The more rings are present in a compound, however, the harder it is for the microbe to degrade it due to its aforementioned hydrophobicity [17]. | ||
==Diverse Specializations of <i>Sphingomonas</i> Species== | ==Diverse Specializations of <i>Sphingomonas</i> Species== | ||
Revision as of 03:31, 26 April 2011
Introduction
The Genus Sphingomonas includes a range of bacterium that are remarkable for their ability to break down polycyclic hydrocarbons. Bacteria in this genus have been detected in a variety of environments, both marine and terrestrial. In recent years, these bacteria have been a focus of study because of their possible applications for bioremediation (the use of biological agents to remove pollutants from the environment). In addition to these helpful functions, however, some strains of this Genus have also been found to cause illnesses.
Characterization of Sphingomonas
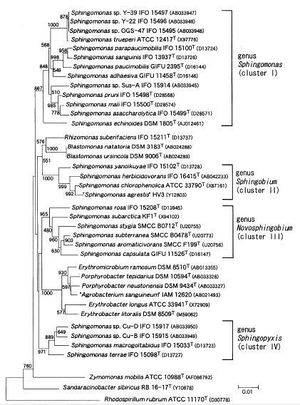
The Genus Sphingomonas includes many bacteria with varied morphological traits. Though initially only a few were none and mostly in a clinical setting, further studies started to reveal many morphologically similar strains. For this reason, in 2001 they were separated into four distinct clusters within the Genus (Figure 2). These clusters, collectively known as the Sphingomonads, are individually known as Sphingomonas, Sphingobium,
Novosphingobium, and Sphingopyxis. There are more than 20 known species that are distributed amongst these strains according to their chemotaxonomic and phenotypic traits, as well as their 16S rRNA gene sequences [22].
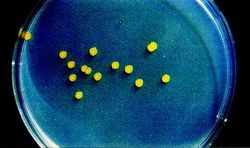
Some shared traits among all strains of Sphingomonas are that they are Gram-negative and possess a bacillus (rod) shape. The bacteria are also strictly aerobic chemoheterophs. Whereas other bacteria utilize lipopolysaccharides in their cell envelopes, Sphingomonas integrate glycosphingolipids. This is the primary distinguishing characteristic separating them from other species in the α-subclass of Proteobacteria. In cultures Sphingomonas colonies typically appear yellow (Figure 3). Finally, Sphingomonas species’ major quinone is characteristically ubiquinone 10 [13].
Identified Species
Sphingomonas: adhaesiva, aerolata, aquatilis, asaccharolytica, aurantiaca, baekryungensis, chungbukensis, cloacae, echinoides, elodea, faeni, koreensis, mali, melonis, natatoria, oligophenolica, parapaucimobilis, paucimobilis, phyllosphaerae, pituitosa, pruni, roseiflava, sanguinis, suberifaciens, taejonensis, trueperi, ursincola, wittichii, xenophaga, yabuuchiae. [2]
Environmental Contamination: The Issue
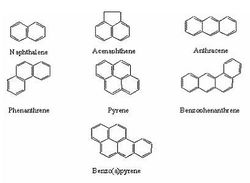
Polycyclic aromatic hydrocarbons (PAHs) are a pressing issue for several reasons, the main being that they are dangerous for both humans and other organisms. PAHs are a major component of many fuels and oils, and therefore enter the environment through vehicle leaks, incorrect disposal of wastes, petroleum spills, and natural seeps. These compounds are extremely stable thanks to a system of double bonds throughout their hydrocarbon rings, and are therefore highly resilient in the environment. An additional problem is that many bacteria cannot degrade chemicals unless they are dissolved in water. Because of their high concentrations of hydrophobic carbon-hydrogen bonds, however, PAHs interact with non-aqueous media and soil organic matter. This often makes them unavailable to degradation and only furthers their persistence. For these various reasons, PAHs have low bioavailability and can be used as carbon sources by only a select few organisms [16].
PAHs are often formed by the incomplete burning of coal, oil, garbage, and other organic substances such as tobacco, wood, and meat. Humans often encounter at least two or more of these substances in the form of soot. Because of their presence in burned meats, they are often ingested. PAHs can also be produced industrially for usage in coal tar, crude oil, creosote and roofing tar. Some other forms are used in medicines, dyes, plastics, and pesticides [2]. Any of these pathways may ultimately contribute to environmental levels, though the greatest quantities enter through oil seeps and spills.
PAHs with as few as three rings are recognized as carcinogens. For residents of the United States, most PAHs are encountered through ingestion, though this can vary depending on diet and lifestyle [1]. In humans, these compounds are known to affect the dermal, immunological, and hepatic systems [2]. PAHs' effects on animals have been tested through oral and dermal application, as well as through inhalation; all of these exposures yield tumors. PAHs have effects on DNA binding and sister chromatid exchange, and can cause chromosomal aberrations and sperm abnormalities. Studies on the effects of PAH on human beings are ongoing [17].
Introduction of microbes capable of bioremediation to an environment is not a simple fix for this problem, however, as several layers to the issue remain: 1) In many parts of the world, much of the year is spent at low temperatures, which are often cold enough to prevent efficient microbial degradation of even ideal substrates. These temperatures may result as simple climate or weather patterns, or they can occur in deep subsurface soil. Either way, they inhibit bioremediation efforts. 2) Lack of oxygen sometimes prevents effective breakdown of complicated materials. In anaerobic conditions substrates show varying degrees of degradability—some can be broken down, albeit slowly, whereas others cannot be broken down at all. In the worst cases, target compounds can only be broken down partway, forming toxic compounds that carry a whole other set of repercussions. 3) Contaminated sites often have low nutrient-balances. 4) Besides the aforementioned low solubility of PAHs in water, any large contaminated area is difficult to completely contact with the bioremediation microbe. 5) Finally, some synthetic contaminants simply cannot be catabolized at the moment. In order to find a manner of degrading them, researchers must first isolate a microbial strain that has evolved to grow on the substance [21].
Breakdown of PAHs
Microbial PAH degradation typically begins with oxidization to cis-dihydrodiols. This ring breakage is energetically costly and is therefore usually the rate-limiting step of the process. This initial oxidation integrates atmospheric oxygen into the substrate (therefore, to utilize this pathway bacteria must be in contact with oxygen). Bacterial dioxygenase enzymes allow them to break the aromatic nucleus and insert the two oxygen atoms. Once the dihydrodiols are produced they will be re-aromatized using a cis-dihydrodiol dehydrogenase to produce a dihydroxylated intermediate. Continued oxidation of the cis-dihydrodiols eventually results in catechols, which can be targeted by other dioxygenases that cause enzyme-induced cleavage of the aromatic ring.
Two pathways of catechol degradation exist: the ortho pathway cleaves the bond between the two carbons carrying the two hydroxyl groups to produce cis,cis-muconic acid, whereas the meta pathway proceeds through cleavage of the bond between a hydroxyl-bearing carbon and the adjacent carbon with a hydroxyl group (Figure 5). The rest of the intermediates in the pathway are continuously processed into components of biosynthesis and energy. In the course of the reaction, carbon dioxide and water are also released. In this manner, the rest of the rings of a PAH will then be attacked; once one ring is reduced to pyruvic acid and carbon dioxide, the process will move onto the adjacent ring. The more rings are present in a compound, however, the harder it is for the microbe to degrade it due to its aforementioned hydrophobicity [17].
Diverse Specializations of Sphingomonas Species
Though Sphingomonas species vary in their ideal habitats, most seem to possess functions useful to bioremediation (sometimes besides their PAH-degrading capacity). Therefore, much research has been focused on finding strains that thrive under conditions where bioremediation may be necessary. Some examples include:
Subsurface Soils
A 1995 study collected bacteria from the United States Southeast Coastal Plain subsurface sediments and subjected them to a battery of tests. Tests of phylogeny using 16S rRNA genes showed that some of the collected strains were closely related to Sphingomonas capsulate. Additionally, it was found that these bacteria contained sphingolipids (characteristic of Sphingomonas). Perhaps most notably, most of these bacteria were able to grow on a variety of aromatic compounds, including both toluene and naphthalene. Though many other gram-negative heterotrophic aerobic bacteria can degrade these compounds, it is rare that one organism can catabolize both. Of the strains collected, one could degrade salicylate and benzoate, two could degrade p-cresol and salicylate, and five strains could degrade flourene, biphenyl, and dibenzothiophene. Though further characterization of these strains is ongoing, they are believed to be members of Genus Sphingomonas, all specializing in catabolism of different compounds at the subsurface level [14].
Aquatic Environments
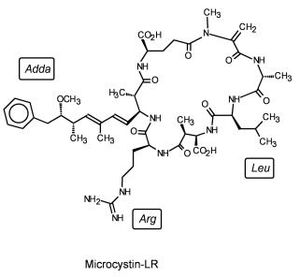
In China a study was performed to assess the abilities of many marine lifeforms to break down microcystin, a common marine contaminant (Figure 6). Among the plants, invertebrates, fish, and bacteria whose abilities were studied was Sphingomonas sp. Sphingomonas was able to catabolize microcystin-LR using three different intracellular hydrolytic enzymes, one of which came to be known as microxystinase. This study represents an avenue for aquatic bioremediation [4].
Cold Soils
At Scott Base in Antarctica, a strain known as Ant 17 was isolated from fuel-contaminated soil. The bacteria grows from temperatures as low as 1oC to 35oC, and can mineralize phenanthrene over a wider range than 24oC. Despite the cold conditions, this strain appeared to be able to metabolize at a rate quicker than should be allowed by mesophilic enzyme kinetics. Indeed, Ant 17 was more effective than Sphingomonas spp. WPO-1 under UV-irradiation and freeze-thaw cycles, indicative of adaptation to the Antarctic environment. Most relevant to bioremediation was the fact that Ant 17 could degrade the aromatic portions of various crude oils, jet fuel, and diesel fuel. Due to its wide aromatic substrate range and ability to catabolize them in low, fluctuating temperatures, Ant 17 appears to be a promising bioremedial microbe for the cleaning of cold, fuel-contaminated environments [11].
Uranium-Contaminated Environments
Often, soil around nuclear reactors and power plants or uranium mines contains uranium (IV) in forms such as [UO2(CO3)2]−2 and [UO2(CO3)3]−4, which can permeate groundwater and lead to health hazards (Figure 7). The Sphingomonas strain BSAR-1 was found to precipitate uranium thanks to an alkaline phosphatase secreted into the outside medium, thus preventing hazards. When the gene encoding this function, dubbed phoK, was inserted into E. coli and overexpressed, the bacteria became powerful uranium precipitators. This recombinant strain precipitated more than 90% of a provided quantity of uranium in less than 2 hrs, whereas the original Sphingomonas degraded this quantity in a little over 7 hrs. This study revealed both certain Sphingomonas species’ facility at bioprecipiation of contaminants, and the possibility that these traits could be transferred to other species to be used in bioremediation [19].
Current Sphingomonas Research for Bioremediation
Improving in situ Bioremediation
Several methods are currently being researched as possible avenues to enhance the rate of bioremediation.
One area of research seeks to enhance the speed at which evolution of local microflora occurs so that it can target contaminating compounds. Typically, this evolution occurs in only a few possible ways. Over time, the microbial population can gain the ability to metabolize the target compound. This method may be time-consuming, however. Another possibility is that local microbiota may encounter microbes immigrating from elsewhere, and therefore obtain the genes needed for biodegradation of contaminants through conjugation and transduction with these foreign bodies. Researchers can promote this transformation by providing the necessary genes to local microbiota in the form of plasmids, with the hope that the plasmids will be taken up as part of these well-adapted species’ genome. Finally, researchers can also simply genetically engineer an organism well-suited to the environment that also has the ability to consume contaminants. This method may be challenging, however, because researchers must be careful to insert suicide functions into the organism to keep its spread controlled, while at the same time producing a strain that will be able to compete against other local life [21].
Another path of current research indicates that the presence of plants may assist in the bioremediation of certain areas. Plant roots penetrate the soil and locally modify conditions in the rhizosphere and mycorrhizosphere. Such plant roots may provide a surface for microbial growth and therefore enhance microbial density and diversity in the soil. Moreover, plant root exudates may represent useful co-substrates for bacteria that improve degradation by inducing useful catabolic pathways. Yet another function of plant roots may be to churn and increase porosity of the soil, which improves aeration. This could be very useful to aerobic microbes such as Sphingomonas, which can only undergo its bioremedial pathways in the presence of atmospheric oxygen [21].
The BP Oil Spill

On April 20, 2010, an explosion shook the BP Deepwater Horizon oil rig, resulting in billions of dollars in losses to British Petroleum, massive trauma to local ecosystems, and the destruction of the livelihood of people whose occupations are based on the USA’s Gulf Coast. It was estimated that 2.5 million gallons of petroleum were leaking into the surrounding ocean per day. This was the worst spill in American history (moreso than the Exxon Valdez spill) and the largest anthropogenic input of petroleum into the environment ever. Though local ecosystems are slowly recovering, research continues to find ways to assist in PAH degradation [7] [3].
Recently, Dr. Waltena Simpson, an associate professor of microbiology at South Carolina State University, has been conducting research on Sphingomonads as a possible remedy for the high quantity of PAHs released by the Deepwater Horizon spill. Her research is focused on a strain of Sphingomonas that was isolated from sludge at a Polish oil refinery once operated by BP. Because these bacteria clearly already possess the ability to degrade PAHs, Dr. Simpson and her partner are seeking to identify the genes that grant the useful catabolic abilities to this strain. After identifying the genes the researchers hope to be able to enhance their effect, and increase the rate at which the microbes catabolize PAHs. This bioengineered strain could then be a useful agent of bioremediaton in the Gulf of Mexico and beyond [15].
Pathological Implications
Despite the useful functions of many Sphingomonas strains, there are a few pathogenic strains that affect a number of organisms.
Sphingomonas paucimobilis: Bacteremia Outbreak in a Hemato/Oncology Unit
Sphingomonas paucimobilis is a rare opportunistic pathogen that has been known to reside on medical equipment. Thus, human infections are often the result of improper handling or production of hospital equipment that is then exposed to ill patients.
In 2007, a bacteremia outbreak in the Gulhane Military Medical Teaching Hospital in Ankara, Turkey, prompted investigators to isolate several strains of the guilty bacteria, four from patients and two from environmental sources, and develop profiles determining the relatedness between the strains, all of which were ultimately classified as Sphingomonas paucimobilis.
Bacteremia, a condition caused by this strain, results when bacteria can be found in the normally sterile environment of the blood, and should be quickly treated (Figure 9). Failure to treat the condition in time may result in spread of the infecting microbes from the original entry site to other portions of the body. In addition to spreading, bacteria may also cause septic shock, which has a relatively high mortality rate [3].
Patients were suspected of infection when their body temperatures rose above 38oC. At this point, blood was drawn and cultured on a medium containing 5% sheep’s blood that would support the suspected bacteria’s growth. Additionally, environmental sources were considered and samples were taken using reliable methods: possible sources included bathtubs, tap water, medical devices, aerators, intravenous fluids, and the hands of health care workers. All of these samples were added to agar plates containing 5% sheep’s blood and incubated at 37oC, which was expected to accommodate growth of the possible pathogen. In total, four cultures were taken from the blood cultures of patients and twenty-eight were taken from the environmental hazards.
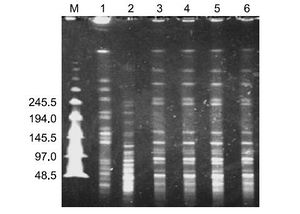
A pulsed-field gel electrophoresis (PFGE) was run after DNA was digested by a DNAse, and the gel was stained with ethidium bromide. If all bacteria were of similar phylogeny, they were expected to produce similar banding patterns. Indeed, this is what was observed for the four blood cultures. The environmental cultures, however, showed unique banding patterns (Figure 10).
Ultimately, the four blood culture bacteria samples were confirmed by the API ID32GN system to be Sphingomonas paucimobilis. The two environmental samples were similarly confirmed, which meant that patients were possibly suffering from nosocomial infections. Although all of the clinical strains had the same genotype, they showed varied resistance to a battery of antibiotic tests. Thus, despite their similar origins, the variability in PFGE and antibiotic susceptibility suggests that different methods must be utilized to combat infections by bacteria from different sources.
Interestingly, despite the fact that bacteremia caused by Gram-negative bacteria often results in high mortality or morbidity rates, no strong correlation between Sphingomonas paucimobilis and death been established. Though the presence of this bacteria may induce bacteremia, treatment with antibiotics is typically sufficient to prevent death. This may be due to the absence of lipopolysaccharides in these bacteria’s cell membranes (characteristic of their genus). The lack of these parts, and therefore their associated endotoxins, may allow patients to effectively recover (the patients in this study, for example, all survived) [18].
Sphingomonas melonis: A Proposed Sphingomonas Affecting Melons
In 2002 researchers examined a phytogenic pathogen that was causing yellow Spanish melon fruits to exhibit brown spots. Two strains of this pathogen were designated DAPP-PG 224 and DAPP-PG 228, respectively. 16S rDNA sequences of the pathogens were isolated and analyzed for phylogeny, which revealed that they were closely related to Sphingomonas mali, Sphingomonas prunii, and Sphingomonas asaccharolytica.
Because the researchers in this study were seeking to prove that melonis belongs in the Genus Sphingomonas, further tests were performed to confirm its identity. The ubiquinones were isolated from the bacteria’s plasma membrane and were tested with reverse-phase TLC. Rf values for the experimental isolates were compared to standard Rf values of known ubiquinones. This method revealed that Q-10 was the most abundant quinone, as would be expected for a Sphingomonas. After Q-10, Q-8 and -9 were the next two highest components.
To ensure that melonis possessed glycosphingolipids, the cellular lipids were collected with chloroform/methanol. The lipids were lysed with NaOH and then run through TLC with glucurosonyl ceramide of S. paucimobilis. Once again, this test yielded a positive result and makes it appear that melonis is indeed a Sphingomonas [12].
Unidentified Sphingomonas Targeting Coral

In June of 1995, a pathogen affecting coral struck the Florida Keys. The disease was characterized by rapid tissue destruction, typically starting at the base of coral bodies and moving upward. Sometimes tissue destruction moved up to 2 cm per day along coral colonies. The disease came to be known as plague type II and is characterized by a line separating the healthy tissue of the coral from the degraded, skeletal portion (Figure 11). Through 1997, Dichocoenia stokesi, the species most affected, experienced up to 38% mortality in the region.
In a study performed during the plague period, syringes were used to collect bacteria from the distinctive line between live and dead tissue on afflicted coral species. This bacteria was cultured and its DNA analyzed, eventually revealing that the most abundant bacteria present was closely related to Sphingomonas. To test whether this bacteria was responsible for plague type II, Dichocoenia stokes coral were placed in tanks on top of bacterial colonies grown in the lab. In all experimental cases all of the exposed coral died of plague, whereas control subjects in separate tanks survived. Besides the microbiological methods, researchers used oxygen-sensitive micro-electrodes to monitor the oxygen concentrations and dynamics in an aerated tank containing either a diseased or healthy coral. Light was shone on the system, but fluctuated on and off.
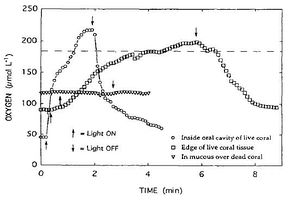
The results of the oxygen dynamics experiment were interesting in that the living portion only had an oxygen concentration of 45.2 μM with the lights off, which is about a quarter of the ambient oxygen concentration of 184 μM. Illumination with photosynthetically active light caused a sharp increase to 217 μM over the course of 2 minutes. Turning the light off caused [O2] to level off after 2.5 minutes. In the diseased tissue a similar trend was observed, though at a slower rate: [O2] in the dark was 90.5 μM, illumination increased it to just above ambient [O2] after five minutes, and return to darkness caused [O2] to decrease to normal levels after 3 minutes. The dead portion of the coral had an [O2] of 117 μM regardless of light (Figure 12).
These values are interesting because they represent the relative respiratory activities of diseased or healthy coral tissue. This is because in the darkness, the only source of oxygen for zooxanthellae (coral tissue) and microorganisms on the coral was the surrounding water. Thus, since the water surrounding healthy coral had the lowest [O2] in the dark, this coral had the highest O2 demand and was consuming the gas. With the lights on photosynthesis was occurring and producing O2, so the concentration spiked. Likewise, diseased tissue showed the same trend but at a lower rate than healthy coral, because metabolic activity (as indicated by oxygen demand) was decreasing as the coral died. Along these lines, it made sense that the dead tissue had the highest surrounding [O2], since there was very little demand for O2 for metabolism. This study revealed that the main pathogen behind the infection was probably the aerobic Sphingomonas, since the disease line moved upwards, where healthy tissue was producing [O2], and away from dead and diseased tissues, which exhibited diminished O2 production [20].
Conclusion
The Genus Sphingomonas is an intriguing example of the variability of lifestyles and morphologies within a single group. These Proteobacteria have gained fame as useful agents of bioremediation, thanks to the assortment of metabolic pathways they exploit and most notably, their ability to break down PAHs. But though the power of these organisms can be harnessed for the good of mankind, within the same Genus exist pathogens such as Sphingomonas paucimobilis that cause infections in human hosts. Future research into this fascinating Genus will reveal more about how we can use some of them as a resource, and how we can defend ourselves against others.
References
[1] "About Coal Tar and Polycyclic Aromatic Hydrocarbons (PAHs)." The Truth About Refined Coal Tar Based Pavement Sealers and PAHs. Web. 25 Apr. 2011. <http://www.truthaboutcoaltar.com/aboutcoaltar.html>.
[2] "Agency for Toxic Substances and Disease Registry - Toxic Substances - Polycyclic Aromatic Hydrocarbons (PAHs)." ATSDR Home. Web. 25 Apr. 2011. <http://www.atsdr.cdc.gov/substances/toxsubstance.asp?toxid=25>.
[3] "Bacteremia Treatment,Bacteremia Treatment India,Cost Bacteremia India." Medical Tourism India,Medical Surgery Tourism India,India Health Tour. Web. 25 Apr. 2011. <http://indiahealthtour.com/treatments/pediatric-surgery/bacteremia-conditions-india.html>.
[4] "Biodegradation of Microcystin in Aquatic Environment--《Ecologic Science》2005年01期." China National Knowledge Infrastructure --- Download Paper. Web. 25 Apr. 2011. <http://en.cnki.com.cn/Article_en/CJFDTOTAL-STKX200501025.htm>.
[5] "Coral Diseases." Bellarmine University. Web. 25 Apr. 2011. <http://cas.bellarmine.edu/tietjen/Ecology/coral_diseases.htm>.http://cas.bellarmine.edu/tietjen/Ecology/coral_diseases.htm
[6] "G.L. Boyer." SUNY-ESF, SUNY College of Environmental Science and Forestry. Web. 25 Apr. 2011. <http://www.esf.edu/biochemistry/boyer.html>.
[7] "Gulf Oil Spill: 10 Horrifying Facts You Never Wanted To Know." Care2 - Largest Online Community for Healthy and Green Living, Human Rights and Animal Welfare. Web. 25 Apr. 2011. <http://www.care2.com/causes/environment/blog/10-most-horrifying-facts-about-the-gulf-oill-spill/>.
[8] "Sphingomonas Wittichii RW1 - Home." JGI Genome Portal - Home. Web. 25 Apr. 2011. <http://genome.jgi-psf.org/sphwi/sphwi.home.html>.
[9] "Uranium (Pollution) - Strange Google Earth Maps." Strange Google Earth Places - Rodsbot.com. Web. 25 Apr. 2011. <http://www.rodsbot.com/?c=3381>.
[10] "Useful Links." Plattsburgh State Faculty and Research Web Sites. Web. 25 Apr. 2011. <http://faculty.plattsburgh.edu/jose.deondarza/MicroWorld/Prokaryotes.htm>.
[11] Baraniecki, C. A., J. Aislabie, and J. M. Foght. 2002. Characterization of Sphingomonas sp. Ant 17, an aromatic hydrocarbon-degrading bacterium isolated from Antarctic soil. Microbial Ecology, Vol 43, No 1, 44-54.
[12] Buonaurio, R., V. M. Stravato, Y. Kosako, N. Fujiwara, T. Naka, K. Kobayashi, C. Cappelli, and E. Yabuuchi. 2002. Sphingomonas melonis sp. nov., a novel pathogen that causes brown spots on yellow Spanish melon fruits. International Journal of Systematic and Evolutionary Microbiology, Vol 52: 2081-2087.
[13] Dworkin, Martin, and Stanley Falkow. "Sphingomonas and Related Genera." The Prokaryotes: a Handbook on the Biology of Bacteria. New York: Springer, 2006. 605-29. Print.
[14] Frederickson, J. K., D. L. Balkwill, G. R. Drake, M. F. Romine, D. B. Ringelberg, and D. C. White. 1995. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Applied and Environmental Microbiology, Vol 61, No 5: 1917-1922.
[15] Gleaton, Dionne. "S.C. State Professor's Research Could Help with Oil Cleanups." Orangeburg South Carolina News. Web. 25 Apr. 2011. <http://www.thetandd.com/news/local/article_1f41762e-8bd4-11df-96e2-001cc4c03286.html>.
[16] Johnsen, Anders R., L. Y. Wick, and H. Harms. 2005. Principles of microbial PAH-degradation in soil. Environmental Pollution Vol 133, Issue 1: 71-84.
[17] Juhasz, Albert L. and R. Naidu. 2000. Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a]pyrene. Internal Biodeterioration & Biodegradation, Vol 45, Issues 1-2: 57-88.
[18] Kilic, Abdullah, Z. Senses, A. E. Kurekei, H. Aydogan, K. Sener, E. Kismet, and A. C. Basustaoglu. 2007. Nosocomial outbreak of sphingomonas paucimobilis bacteremia in a hemato/oncology unit. Japanese Journal of Infectious Diseases, Vol 60: 394-396.
[19] Nilgiriwala, Kayzad S., A. Alahari, A. S. Rao, and S. K. Apte. 2008. Cloning and overexpression of alkaline phosphatase phoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Applied and Environmental Microbiology, Vol 74, No 17: 5516-5523.
[20] Richardson, Laurie L., W. M. Goldberg, R. G. Carlton, and J. C. Halas. 1998. Coral disease outbreak in the Florida Keys: Plague Type II. International Journal of Tropical Biology and Conservation, Vol 46(5): 187-198.
[21] Romantschuk, M., I. Sarand, T. Petanen, R. Peltola, M. Jonsson-Vihanne, T. Koivula, K. Yrjälä, and K. Haahtela. 2000. Means to improve the effect of in situ bioremediation of contaminated soil: an overview of novel approaches. Environmental Pollution, Vol 107, Issue 2: 179-185.
[22] Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu strict and three new genera, Sphingobium, Novosphingobium, and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. International Journal of Systematic and Evolutionary Microbiology, Vol 51: 1405-1417.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2010, Kenyon College.