Spiroplasma melliferum: Difference between revisions
| Line 48: | Line 48: | ||
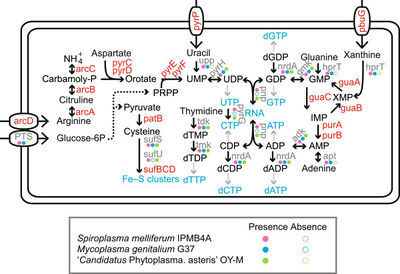
[[File:Metabolic_Pathways.jpeg|thumb|right|400px|The Metabolic Pathways of <I>Spiroplasma melliferum</I>]] | [[File:Metabolic_Pathways.jpeg|thumb|right|400px|The Metabolic Pathways of <I>Spiroplasma melliferum</I>]] | ||
<I>Spiroplasma melliferum</I> ferments glucose and hydrolyzes arginine (1). It also possesses all the genes required for glycolysis to convert glucose-6-phophate into pyruvate which is used for the production of cysteine (1). | <I>Spiroplasma melliferum</I> ferments glucose and hydrolyzes arginine (1). It also possesses all the genes required for glycolysis to convert glucose-6-phophate into pyruvate which is used for the production of cysteine (1). | ||
==Ecology== | ==Ecology== | ||
Revision as of 20:32, 12 January 2014
A Microbial Biorealm page on Spiroplasma melliferum
Classification
Higher Order Taxa:
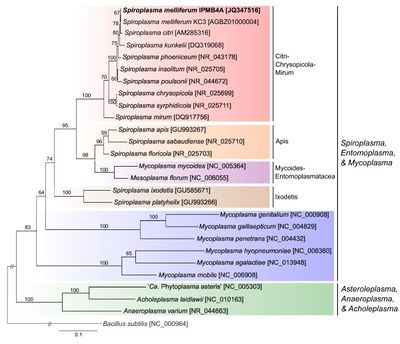
Class: Mollicutes
Order: Mycoplasmatales
Family: Spiroplasmataceae
Genus: Spiroplasma
Species: melliferum
NCBI Accession #: JQ347516
Description and Significance
Genome Structure
The whole-genome shotgun sequencing of S. Melliferum IPMB4A produced a draft assembly that was ~1.1 Mb in size and covered ~80% of the chromosome (1).
Cell and Colony Structure
Spiroplasma melliferum is a helical, motile bacteria without cell walls (1). Spiroplasma melliferum has a classic fried-egg shaped colony morphology.
Metabolism
Spiroplasma melliferum ferments glucose and hydrolyzes arginine (1). It also possesses all the genes required for glycolysis to convert glucose-6-phophate into pyruvate which is used for the production of cysteine (1).
Ecology
Endosymbiotic inhabitants.
Pathology
While most of the Spiroplasma species appeared to be harmless commensals of insects, a small number of species have evolved pathogenicity toward various arthropods and plants (1).
Host: infects the honeybee.
References
1) Wen-Sui Lo, Ling-Ling Chen, Wan-Chia Chung, Gail Gasparich, Chih-Horng Kuo. (2013) Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics. 9 January 2014. http://www.biomedcentral.com/1471-2164/14/22
2) Clark, T. B., R. F. Whitcomb, J. G. Tully, C. Mouches, C. Saillard, J. M. Bove, H. Wroblewski, P. Carle, D. L. Rose, R.B. Henegar, and D. L. Williamson. (1985) Spiroplasma melliferum, a New Species from the Honeybee (Apis mellifera). International Journal of Systematic Bacteriology, Vol. 35, No. 3, 296-308.
Edited by Brittany Demmons, student of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio