Staphylococcus epidermidis: Difference between revisions
| Line 18: | Line 18: | ||
==Description and significance== | ==Description and significance== | ||
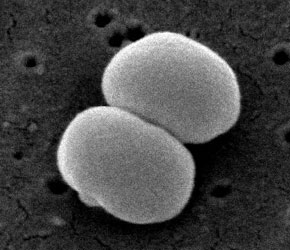
Staphylococcus epidermidis is a gram-positive and coagulase-negative staphylococci (4). It typically lives on the human skin and mucosa and the most common infections on catheters and implants (5). S. epidermidis is one of five most common organisms that cause noscomial infections due to the increase in usage of biomaterials in the clinical environment (8). The nosocomial pathogen causes infections on prosthetic valves, cerebrospinal fluid shunts, joint prosthesis vascular prostheses, valves, and in postoperative wounds and the urinary tract. It is also the most freqent organism found in the blood of bone marrow transplant patients and on central venous catheters for patients of total parenteral nutition (4). A controversial issue on S. epidermidis argues whether or not all strains can equally cause disease and if a specific genotype determines a specific disease (6). | |||
S. epidermidis strain from health adult nares show that there are many kinds of the organism in each individual (9). A common S. epidermidis strain is RP62a (ATCC 35984) was isolated in Memphis, Tennessee during 1979-1980 in a wide spread of intravascular catheter sepsis. RP62a is a strain that produces slime, grows collectively and forms biofilm. The treatment of S. epidermidis infections caused by biofilm with antibiotics is often ineffective and results in removal of the implants (5). Another strain is S. epidermidis ATCC 12228 which does not produce biofilm (7). | |||
The ability to produce slime allows the pathogen to stay on biomaterials. Different strains of S. epidermidis can be distinguished by whether or not they produce slime. In clinical laboratories, S. epidermidis infections are studied by the use of antibiograms, biotyping, plasmid profiles, and slime production; whereas in research, phagetyping, serotyping, restrict enzyme analysis and DNA-DNA hybridization are used (4). | |||
==Genome structure== | ==Genome structure== | ||
Revision as of 18:36, 29 August 2007
A Microbial Biorealm page on the genus Staphylococcus epidermidis
Classification
Higher order taxa
Bacteria; Firmicutes; Bacillales; Staphylococcus (1)
Species
Genus Species: Staphylococcus epidermidis
Other Names: Micrococcus epidermidis, Albococcus epidermidis, Staphylococcus epidermidis albus (2)
|
NCBI: Taxonomy |
Description and significance
Staphylococcus epidermidis is a gram-positive and coagulase-negative staphylococci (4). It typically lives on the human skin and mucosa and the most common infections on catheters and implants (5). S. epidermidis is one of five most common organisms that cause noscomial infections due to the increase in usage of biomaterials in the clinical environment (8). The nosocomial pathogen causes infections on prosthetic valves, cerebrospinal fluid shunts, joint prosthesis vascular prostheses, valves, and in postoperative wounds and the urinary tract. It is also the most freqent organism found in the blood of bone marrow transplant patients and on central venous catheters for patients of total parenteral nutition (4). A controversial issue on S. epidermidis argues whether or not all strains can equally cause disease and if a specific genotype determines a specific disease (6).
S. epidermidis strain from health adult nares show that there are many kinds of the organism in each individual (9). A common S. epidermidis strain is RP62a (ATCC 35984) was isolated in Memphis, Tennessee during 1979-1980 in a wide spread of intravascular catheter sepsis. RP62a is a strain that produces slime, grows collectively and forms biofilm. The treatment of S. epidermidis infections caused by biofilm with antibiotics is often ineffective and results in removal of the implants (5). Another strain is S. epidermidis ATCC 12228 which does not produce biofilm (7).
The ability to produce slime allows the pathogen to stay on biomaterials. Different strains of S. epidermidis can be distinguished by whether or not they produce slime. In clinical laboratories, S. epidermidis infections are studied by the use of antibiograms, biotyping, plasmid profiles, and slime production; whereas in research, phagetyping, serotyping, restrict enzyme analysis and DNA-DNA hybridization are used (4).
Genome structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence? Does it have any plasmids? Are they important to the organism's lifestyle?
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
Edited by Angela Cheng, student of Rachel Larsen at UCSD
