Synechococcus and Biofuel: Difference between revisions
Cyoung9350 (talk | contribs) |
Cyoung9350 (talk | contribs) |
||
| Line 26: | Line 26: | ||
===Hydrogen Fuel=== | ===Hydrogen Fuel=== | ||
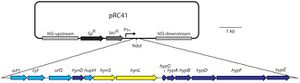
Hydrogen can be produced by hydrogenase or nitrogenase enzymes in cyanobacteria. Nitrogenase system is only present in certain species of cyanobacteria. While it does produce hydrogen, nitrogenase is highly sensitive to oxygen, making it ineffective as a source of efficient hydrogen production. [http://web.b.ebscohost.com/ehost/pdfviewer/pdfviewer?sid=737ba83b-6fd6-469a-914b-83241b63abf9%40sessionmgr115&vid=2&hid=125[11]] Hydrogenase enzymes can be divided into two classes: uptake or bidirectional. Uptake hydrogenases are only able to catalyze reactions from right to left whereas bidirectional hydrogenases are able to catalyze reactions in both directions. It is the bidirectional pathway that is of greater interest as the uptake hydrgoenases mainly recycle hydrogen as a byproduct of nitrogen-fixation. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3102683/[12]] As hydrogenase enzymes are also sensitive to the presence of oxygen as well, there have been several approaches taken to counteract this problem. Asada et. al (1999) were one of the first to illustrate that hydrogen production in S. elongatus PCC7942 was possible. The paper genetically modified S. elongatus PCC7942 to contain a clostridial hydrogenase gene. The transformation was successful and the modified bacterial strains were able to produce hydrogen in the presence of light. This replacement of nitrogenase in heterocysts with hydrogenase increases the efficiency of the oxygen and hydrogen evolution conducted by this bacterial species. The ability of S. elongatus to produce hydrogen also has implications for hydrogen fuel as it provides a source for production. [http://www.ncbi.nlm.nih.gov/pubmed/10684972[13]] A more recent approach has been genetically modifying <i>Synechococcus</i> to express oxygen-tolerant [NiFe] hydrogenases. This experiment, conducted by Weyman et. al in 2011, managed to express stable hydrogenase genes (HynSL) from two different bacterium (<i>Alteromonas macleodii</i> and <i>Thiocapsa roseopersicina</i>) in <i>Synechococcus elongatus | [[Image:Hydrogenase Expression.jpg|thumb|300px|right|Diagram of pRC41 used for expression of AltDE hydrogenase HynSL in S. elongatus. Weyman et. al 2011[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3102683/[12]] | ||
While expression was successfully achieved, further experimentation is needed to increase expression such that hydrogen may be produced during photosynthesis in cyanobacteria such as <i>Synechococcus</i>. | |||
Hydrogen can be produced by hydrogenase or nitrogenase enzymes in cyanobacteria. Nitrogenase system is only present in certain species of cyanobacteria. While it does produce hydrogen, nitrogenase is highly sensitive to oxygen, making it ineffective as a source of efficient hydrogen production. [http://web.b.ebscohost.com/ehost/pdfviewer/pdfviewer?sid=737ba83b-6fd6-469a-914b-83241b63abf9%40sessionmgr115&vid=2&hid=125[11]] Hydrogenase enzymes can be divided into two classes: uptake or bidirectional. Uptake hydrogenases are only able to catalyze reactions from right to left whereas bidirectional hydrogenases are able to catalyze reactions in both directions. It is the bidirectional pathway that is of greater interest as the uptake hydrgoenases mainly recycle hydrogen as a byproduct of nitrogen-fixation. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3102683/[12]] As hydrogenase enzymes are also sensitive to the presence of oxygen as well, there have been several approaches taken to counteract this problem. Asada et. al (1999) were one of the first to illustrate that hydrogen production in S. elongatus PCC7942 was possible. The paper genetically modified S. elongatus PCC7942 to contain a clostridial hydrogenase gene. The transformation was successful and the modified bacterial strains were able to produce hydrogen in the presence of light. This replacement of nitrogenase in heterocysts with hydrogenase increases the efficiency of the oxygen and hydrogen evolution conducted by this bacterial species. The ability of S. elongatus to produce hydrogen also has implications for hydrogen fuel as it provides a source for production. [http://www.ncbi.nlm.nih.gov/pubmed/10684972[13]] A more recent approach has been genetically modifying <i>Synechococcus</i> to express oxygen-tolerant [NiFe] hydrogenases. This experiment, conducted by Weyman et. al in 2011, managed to express stable hydrogenase genes (HynSL) from two different bacterium (<i>Alteromonas macleodii</i> and <i>Thiocapsa roseopersicina</i>) in <i>Synechococcus elongatus</i>. | |||
===Lipid-based Biodiesel=== | ===Lipid-based Biodiesel=== | ||
Revision as of 09:58, 15 April 2014
Synechococcus is a genus of unicellular marine cyanobacteria that are found in both freshwater and marine water environments. They comprise a vast majority of picoplanktonic marine cyanobacteria. [1] Species in this genus are capable of photoheterotrophic, chemoheterotrophic, photoautotrophic, and even nitrogen fixation. [2] The genomes of several species of Synechococcus have been fully sequenced, most notably Synechococcus elongatus strain PCC 7942 and species strain WH8102. Due to the diversity in its metabolism as well as a genome that can be readily manipulated, Synechococcus holds potential for biofuel production via its metabolic pathways
Introduction
Classification
Taxonomy
Bacteria; Cyanobacteria; Chroococcales; Synechococcus [3]
Cell Structure
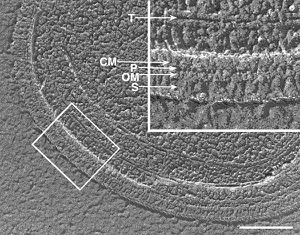
Synechococcus are coccoid shaped bacteria that range in size from 0.2-2 µm. They are classified as Gram-negative cells since the cell envelopes of these cyanobacteria structurally resemble those of Gram-negative bacteria, consisting of a cell membrane, peptidoglycan layer, outer membrane, surface layer, and sometimes additional structures. [4] [5] Synechococci species are both motile and non-motile, although many motile species appear to accomplish their movement without the aid of a flagella. Cells are thought to undergo locomotion through a gilding fashion, using filamentous protrusions (pili or spicules) to make contact with cell surfaces and move in a direction parallel to the filament (directional motility). [7] Swimming motility is believed to be accomplished by surface waves generated by filamentous protrusions. It is proposed that the waves are generated by a motor force embedded in the cell membrane, although the exact mechanism is unclear. [6]
Envrionment & Lifecycle
Synechococcus are found in both the coastal marine environment as well as the pelagic zone (open sea). The environment in which these cyanobacteria are found in is important to regulation of their cell cycles, as factors such as light-dark cycle can affect Synechococcus cell cycles. Gaps in DNA synthesis are found between groups of cells that were exposed to light and exposed to dark. It has been determined that the Synechococcus cell cycle is light-dependent. [8] Synechococcus has 2 modes of cell cycle regulation. The first mode was consistent with the slow-growth model of the prokaryotic cell cycle. The second mode involved unsynchronized chromosome replication, with variability in chromosome copies amongst cells. The second mode serves as a poor estimator of colony growth rate due to the variation in distribution of DNA in the cells adding a probalistic component. [9] Synechococcus undergoes all phases of the cell cycle, although the duration of these phases is dependent upon growth rate and environment. Environmental conditions such as presence of nitrogen can affect the doubling time of the colonies as well as the duration of each phase of the cell cycle (S, G1, G2). Liu et al. found in 1999 that when grown under nitrogen-limited conditions, "(1) the fraction of cells in the S and G2 phases relatively constant, and (2) the cellular carbon, nitrogen and pigment content of this strain and their ratios vary with growth rate." [10] This study illustrated the relationship between physiological characteristics and the cell cycle of Synechococcus, providing greater insight as to better predicting the growth rate of these species.
Metabolism
Carbon Metabolism
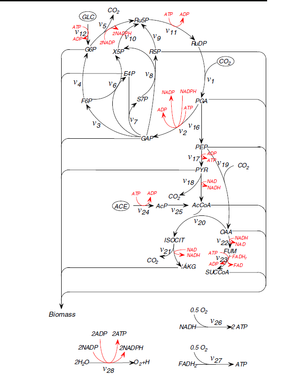
The main Synechococcus species of focus, strain PCC 7942 is considered an obligate photoautotroph. Due to this, it is difficult to cultivate Synechococcus as it undergoes photoinhibtion at high light intensities; at low light intensities there isn’t sufficient energy for cultivation. However, studies have shown that although Synechococcus is an obligate photoautotroph, it is able to incorporate organic carbon sources as a part of its metabolic processes. Synechococcus exhibits several pathways for carbon metabolism. Its main pathway for carbon metabolism under photoautotrophic conditions involves glycolysis, pentose phosphate shunt, incomplete TCA cycle, and photophosphorylation (Yan et al. 2009)[18]. Under mixotrophic conditions involving either glucose or acetate, these compounds are incorporated as organic carbon compounds. Yan et al. showed in 2010 that glucose is metabolized through the main pathway and the CO2 released through respiration and fixed by photosynthesis (Calvin cycle)[19]. ATP is produced via three metabolic pathways: substrate phosphorylation, oxidative phosphorylation, and photophosphorylation. Substrate phosphorylation is the production of ATP by the donation of a phosphoryl group (PO3) from the metabolites in all reaction pathways. In oxidative phosphorylation, which is carried out using an electron transport chain, ATP is generated using either NADH or FADH with O2 as the terminal electron acceptor. Photophosphorylation uses light energy to convert ADP to ATP through photosynthetic reactions.
Nitrogen Metabolism
It has been found that several marine Synechococcus species utilize cyanase to convert cyanate to carbon dioxide and ammonia: NCO- + HCO3- + 2H+ -> 2CO2 + NH3. Cyanase is encoded for by the gene cynS, which is found in many a variety of organisms, including cyanobacteria, fungi, plants, etc. It was proposed by Kammenaya and Post in 2010 that the purpose of cyanase was related to the “detoxification of cyanate that resulted from intracellular urea or carbamoyl phosphate decomposition.”[20] The team found that the metabolism of exogenous cyanate by Synechococcus species was a light-dependent process that did not require previous exposure to cyanate. Furthermore, the group was able to characterize a novel cyanase gene that was found to be highly conserved among seven marine Synechoccocus species known as cynH. This gene was found to be a homolog of cynS, as it contained three completely conserved amino acid residues identical to that of cynS. Also, secondary structure predictions for the C-terminal domain of cynH is in agreement with that of cynS. cynS has been found to play a role in cyanate detoxification, but was not found to be involved in utilization functions of cyanase (cyanate acquisition). The characterization of the cynH gene may highlight the different roles that each gene may play in nitrogen metabolism (cynS) and nitrogen assimilation (cynH). However, further research is needed before the exact nature of cynH is confirmed, as well as the roles of these two cyanase genes in nitrogen metabolism of marine Synechococcus species.
Biofuel Potential and Application
Hydrogen Fuel
While expression was successfully achieved, further experimentation is needed to increase expression such that hydrogen may be produced during photosynthesis in cyanobacteria such as Synechococcus.
Hydrogen can be produced by hydrogenase or nitrogenase enzymes in cyanobacteria. Nitrogenase system is only present in certain species of cyanobacteria. While it does produce hydrogen, nitrogenase is highly sensitive to oxygen, making it ineffective as a source of efficient hydrogen production. [11] Hydrogenase enzymes can be divided into two classes: uptake or bidirectional. Uptake hydrogenases are only able to catalyze reactions from right to left whereas bidirectional hydrogenases are able to catalyze reactions in both directions. It is the bidirectional pathway that is of greater interest as the uptake hydrgoenases mainly recycle hydrogen as a byproduct of nitrogen-fixation. [12] As hydrogenase enzymes are also sensitive to the presence of oxygen as well, there have been several approaches taken to counteract this problem. Asada et. al (1999) were one of the first to illustrate that hydrogen production in S. elongatus PCC7942 was possible. The paper genetically modified S. elongatus PCC7942 to contain a clostridial hydrogenase gene. The transformation was successful and the modified bacterial strains were able to produce hydrogen in the presence of light. This replacement of nitrogenase in heterocysts with hydrogenase increases the efficiency of the oxygen and hydrogen evolution conducted by this bacterial species. The ability of S. elongatus to produce hydrogen also has implications for hydrogen fuel as it provides a source for production. [13] A more recent approach has been genetically modifying Synechococcus to express oxygen-tolerant [NiFe] hydrogenases. This experiment, conducted by Weyman et. al in 2011, managed to express stable hydrogenase genes (HynSL) from two different bacterium (Alteromonas macleodii and Thiocapsa roseopersicina) in Synechococcus elongatus.
Lipid-based Biodiesel
Perhaps the most promising biofuel alternative that Synechococcus is utilized in producing is biodiesel. The cyanobacterium is capable of producing precursors for biodiesel production in the form of fatty acids, or by converting carbon dioxide into energy dense fuel molecules. Synechococcus has a distinct advantage to other cyanobacteria for fatty acid production as it doesn’t contain a polyhydroxybutarate (PHB) pathway. When compared to two other cyanobacteria (Cyanobacterium aponinum and Phormidium) Synechococcus had the highest yield of lipid components, at 42.8%. [15] This pathway has been known to compete with the free fatty acid (FFA) synthesis pathway for available carbon. In a ground-breaking study published by Ann Ruffing and Howland Jones in 2012, the paper not only managed to achieve free fatty acid synthesis (FFA) via genetic manipulation in Synechococcus elongatus PCC 7942, but was also able to identify specific changes in the physiology and metabolism of Synechococcus that limit FFA synthesis. Ruffing and Jones aimed to increase FFA production by creating a gene knockout of a FFA recycling enyme (FFA-recycling acyl-ACP synthetase) and expressing a thioesterase for the increased release of FFA. While, increased production of FFA was noted, the yields were not sufficient for large-scale production. Furthermore, the increased production compromised cell health in a variety of ways. A main concern was the toxicity of unsaturated FFA, which can oxidize into toxic products (reactive oxygen species). Effects of this toxicity include reduced cell growth, photosynthetic yield, and photosynthetic enzyme production. Physiological changes in the cells such as changes in membrane lipid composition, membrane integrity and structure, and the activity of membrane-bound enzymes involved in photosynthesis. However, some of these concerns can be combated by metabolic engineering. For example, the formation of reactive oxygen species (ROS) can be remedied by genetically engineering the production of ROS degrading enzymes in Synechococcus cells. [16]
An alternative method to the production of lipid biofuels is the direct conversion of CO2 into alcohol-based biofuels using genetically engineered Synechococcus cells. The first successful photosynthetic production of 2-methyl-1-butanol (2MB), an energy-dense fuel molecule, from CO2 in genetically engineered cyanobacterium Synechococcus elongatus PCC7942 was achieved in 2012 by Shen and Liao. [17] 2MB is a higher-chain alcohol, specifically belonging to a group of pentanols, meaning that it is more desirable as a fuel molecule due to its higher energy density and lower hygroscopicity. The success of 2MB production is promising for the future of biofuel production. 2MB serves multiple industrial purposes other than being a future fuel molecule. It is also used as an industrial solvent, in chemical engineering, and as a flavor component in foods and beverages. Furthermore, this method is sustainable, as CO2 will be recycled by the photosynthetic Synechococcus bacteria and incorporated in the production of 2MB.
Electrical Fuel Cells
Issues in Current Development
Potential for Mass Production
Future Directions
Further Reading
[Sample link] Ebola Hemorrhagic Fever—Centers for Disease Control and Prevention, Special Pathogens Branch
References
Edited by (Caitlyn Young), a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
