Syntrophobacter: Difference between revisions
No edit summary |
|||
| (26 intermediate revisions by the same user not shown) | |||
| Line 18: | Line 18: | ||
=Description= | =Description= | ||
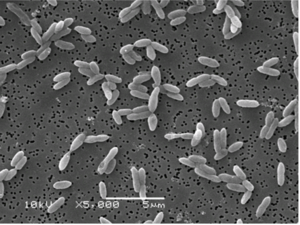
''Syntrophobacter'' is an egg or rod shaped, gram-negative, non-spore-forming and non-motile bacterium approximately 1.0-1.3 x 1.8-2.2 micrometers. ''S. fulfatireducens'' grows between 20 and 48 degrees C and has a doubling time between 52 and 55 hours at 37C (5). | |||
The first species named species of ''Syntrophobacter'' (''S. wolinii'') was discovered in 1980 by Boone and Bryant (1). Since then, three other species of ''Syntrophobacter'' have been described. All members of this genus anaerobically degrade propionate to acetate in the presence of methanogens. | |||
While it was initially thought that ''Syntrophobacter'' requires coculture of at least one methanogen, at least two species (''S. pfennigii'' and ''S. wolinii'') have been grown in pure culture (2,3). | |||
''Syntrophobacter'' is often found in wastewater treatment facilities and is useful for further degrading organic compounds from propionate and lactate to acetate. It is generally found closely associated with methanogens which further process organic material from wastewater. | |||
''S. fumarodoxans'' degrades propionate via the methylmalonyl-CoA pathway when in co-culture with a methanogen. In pure culture, it utilizes sulfate or fumarate as an electron acceptor. In both cases the end products are acetate and carbon dioxide(4). | |||
A genome sequence of ''S. fumarodoxans'' was obtained in 2012. It's genome is nearly 5Mbp long on a single circular genome. It has a 59.95% GC content (4). | |||
[[File:s_fumaroxidans.png|300px|thumb|right|SEM of ''S. fumaroxidans'' [1F] ]] | |||
=References= | =References= | ||
1. Boone DR, Bryant MP. Propionate-Degrading Bacterium, ''Syntrophobacter wolinii'' sp. nov. gen. nov., from Methanogenic Ecosystems. Applied and Environmental Microbiology. 1980;40(3):626-632. | |||
2. Wallrabenstein C., Hauschild E., Schink B.. ''Syntrophobacter pfennigii'' sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Archives of Microbiology. 1995;164(5):346-352. | |||
3. Wallrabenstein, C., Hauschild, E. and Schink, B. (1994), Pure culture and cytological properties of ''Syntriphobacter wolini''. FEMS Microbiology Letters, 123: 249–254 | |||
4. Plugge, C., Henstra, A., Worm, P., Swarts, D., Paultisch-Fuchs, A., Scholten, J., Lykidis, A., Lapidus, A., Goltsman, E., Kim, E., Mcdonald, E., Rohlin, L., Crable, B., Gunsalus, R., Stams, A., Mcinerney, M.. Complete genome sequence of ''Syntrophobacter fumaroxidans'' strain (MPOBT). Standards in Genomic Sciences, North America, 7, oct. 2012 | |||
5. Chen, S., Liu, X., and Dong, X.. ''Syntrophobacter sulfatireducens'' sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. International Journal of Systematic and Evolutionary Microbiology. 2005;55(3):1319-1324 | |||
==Author== | ==Author== | ||
Page authored by | Page authored by Troy Mellen, student of Prof. Katherine Mcmahon at University of Wisconsin - Madison. | ||
<!-- Do not remove this line-->[[Category:Pages edited by students of Katherine Mcmahon at University of Wisconsin - Madison]] | <!-- Do not remove this line-->[[Category:Pages edited by students of Katherine Mcmahon at University of Wisconsin - Madison]] | ||
Latest revision as of 23:23, 13 May 2015
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Deltaproteobacteria
Order: Syntrophobacterales
Family: Syntrophobacteraceae
Genus: Syntrophobacter
Species: S. fumaroxidans; S. pfennigii; S. sulfatireducens; S. wolinii
Description
Syntrophobacter is an egg or rod shaped, gram-negative, non-spore-forming and non-motile bacterium approximately 1.0-1.3 x 1.8-2.2 micrometers. S. fulfatireducens grows between 20 and 48 degrees C and has a doubling time between 52 and 55 hours at 37C (5).
The first species named species of Syntrophobacter (S. wolinii) was discovered in 1980 by Boone and Bryant (1). Since then, three other species of Syntrophobacter have been described. All members of this genus anaerobically degrade propionate to acetate in the presence of methanogens.
While it was initially thought that Syntrophobacter requires coculture of at least one methanogen, at least two species (S. pfennigii and S. wolinii) have been grown in pure culture (2,3).
Syntrophobacter is often found in wastewater treatment facilities and is useful for further degrading organic compounds from propionate and lactate to acetate. It is generally found closely associated with methanogens which further process organic material from wastewater.
S. fumarodoxans degrades propionate via the methylmalonyl-CoA pathway when in co-culture with a methanogen. In pure culture, it utilizes sulfate or fumarate as an electron acceptor. In both cases the end products are acetate and carbon dioxide(4).
A genome sequence of S. fumarodoxans was obtained in 2012. It's genome is nearly 5Mbp long on a single circular genome. It has a 59.95% GC content (4).
References
1. Boone DR, Bryant MP. Propionate-Degrading Bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from Methanogenic Ecosystems. Applied and Environmental Microbiology. 1980;40(3):626-632.
2. Wallrabenstein C., Hauschild E., Schink B.. Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Archives of Microbiology. 1995;164(5):346-352.
3. Wallrabenstein, C., Hauschild, E. and Schink, B. (1994), Pure culture and cytological properties of Syntriphobacter wolini. FEMS Microbiology Letters, 123: 249–254
4. Plugge, C., Henstra, A., Worm, P., Swarts, D., Paultisch-Fuchs, A., Scholten, J., Lykidis, A., Lapidus, A., Goltsman, E., Kim, E., Mcdonald, E., Rohlin, L., Crable, B., Gunsalus, R., Stams, A., Mcinerney, M.. Complete genome sequence of Syntrophobacter fumaroxidans strain (MPOBT). Standards in Genomic Sciences, North America, 7, oct. 2012
5. Chen, S., Liu, X., and Dong, X.. Syntrophobacter sulfatireducens sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. International Journal of Systematic and Evolutionary Microbiology. 2005;55(3):1319-1324
Author
Page authored by Troy Mellen, student of Prof. Katherine Mcmahon at University of Wisconsin - Madison.