The Role of Viral Proteins in Epstein-Barr Virus Induced Disease
Introduction
The Epstein-Barr Virus (EBV) is a common human herpes virus that can cause both infectious mononucleosis and lymphoproliferative disease. EBV is unique in that it infects about 95% of the adult population between 35-40 years old in the U.S. [1]. In most cases, EBV infectious cause no symptoms, particularly if the patient is infected as a child [1,8]. Infection during adolescence are associated with mononucleosis about half of the time [1]. EBV is associated with cancers such as Burkitt’s Lymphoma and nasopharyngeal carcinoma [1,2]. The virus is capable of infecting host B-cells, and primarily proliferates via a non-lytic mechanism [2]. During this latent process, virus-derived nuclear proteins (EBNAs) and membrane proteins (LMPs) are expressed by infected host cells [2]. An advancing area of research is aimed at understanding how viral proteins may play a role in lymphoproliferative disease. LMP-1 is one of these viral membranous proteins that may induce indefinite, tumorigenic replication in infected B-cells [2,3]. In a sense, the LMP-1 acts to "transform" these B-cells into an immortalized, proliferating line. While EBV infections usually only cause mild symptoms, attempts to develop treatments and antivirals have generally been unsuccessful [1]. It is particularly difficult to control spread between hosts as chronic infections may reactivate, allowing symptom-free carriers to continue to transmit the virus years after initial infection [1]. While the latent life cycle appears to be the primary contributor to the incidence of lymphoproliferative disease in EBV infection, the lytic system makes a significant contribution. Lytic mechanisms are essential for horizontal spread of the virus, and several lytic cycle associated mechanisms contribute to both B-cell proliferation or success rates of infection [2,9,11]. In a sense, lytic mechanisms are essential for primary infection of humans, but once established in the body, latent mechanisms prevail in maintaining an infection in a host [2,22]. Due to EBV's putative role in carcinogenesis and the limited success in development of antivirals to combat infection, it is essential to develop a deeper understanding of the mechanisms by which the virus alters the character of host cells.
Virus Classification and Genome
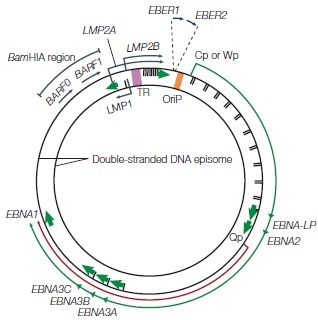
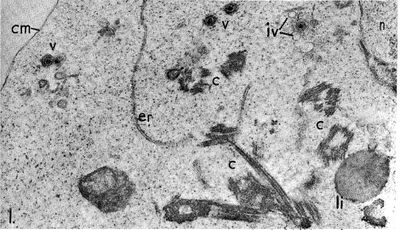
The Epstein-Barr virus is a Baltimore Class I virus of the family Herpesviridae. It is a gammaretrovirus also designated as Human Herpes Virus 4 (HHV-4). M.A. Epstein and Yvonne Barr were the first identify EBV in tumor tissue associated with Burkitt's Lymphoma. The researchers used electron microscopy to determine that these viral particles were very similar in structure to Herpes simplex virions (Figure 1)[4]. The genome of EBV is composed of double-stranded DNA and is 172,282 base-pairs long [5]. This relatively long genome is characteristic of Herpes viruses; Herpes simplex has a 152-kilobase genome [6]. The open reading frames (ORFs) or EBV are generally broken up into separate lytic and latent sections [7]. While most of the viral genes encode proteins, some of the latent genes remain untranslated (EBERs: ENV encoded RNAs) and several micro-RNAs are coded for [7]. The relativity large dsDNA genome of the Epstein-Barr virus exhibits separation of ORF sections coding for genes associated with its two different replicative strategies.During latent infection, the EBV genome exists in a circularized form localized in the host cell nucleus [2]. The reading frames for the LMP and EBNA proteins are clustered separately within the episome (Figure 2).
EBV Virion Structure
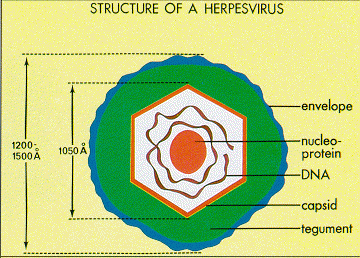
Epstein and Barr observed that this lymphoma-associated virus was about 20% smaller than typical Herpes simplex virions [4]. Similar to other Herpesviruses, the innermost part of the EBV virion consists of DNA wrapped around a central nucleo-protein core (Figure 3) [8]. The core of the virion is surrounded by a nucleocapsid, a layer of protein tegument, and finally and outer envelope with spike glycoproteins [8]. Similar to many other viruses, many of these glycoproteins are vital for host-cell entry mechanisms. Infected host cells release EBV virions exclusively during the lytic cycle [7].
Lytic Life Cycle
While the EBV lytic life cycle is more rarely observed than the latent mode of replication, it is particularly important as it is the only way that the virus may make virions and be transferred horizontally between hosts (or cells) [8,9]. Immunosuppresive diseases like AIDS typically show increased free virion levels in the blood, a marker of increased lytic activity [9]. While virions are often found in the saliva of infected hosts, little to no lytic-infected cells are typically detected in the body [9]. Human cytotoxic T-cells are particularly adept at recognizing and destroying lytically infected cells expressing certain early stage lytic genes [10]. Hence, lytic activity appears to drive EBV spread in human populations. However, the latent replicative cycle is favored under normal conditions within a host, possibly as a means to evade host immune responses. Alpha and Beta hepesviruses have elaborate mechanisms for lytic gene concealment from the host immune system while EBV has few mechanisms to prevent immunomediated destruction of lytic-infected cells [10]. For example, Herpes simplex may be capable of inhibition of host Major Histocombatibility Complex, which reduces B-cell antigen presentation and recognition by cytotoxic T-cells [10]. In contrast to other herpesviruses, gammaherpesviruses like EBV are capable of proliferation via clonal replication within host B-cells [10]. Since EBV can replicate effectively via this latent system, there is likely a reduced selective pressure for elaborate immune-avoidance mechanisms in the lytic cycle. Said another way, in contrast to other herpes viruses, where lytic replication is essential for continued, proliferative infection within a host, lytic replication in EBV takes on a specific "niche role" as a mediator of host-host transmission.
While latent activity typically is the primary contributor to the development of lymphoproliferative diesease (LPD) in EBV infected individuals, lytic-incompetent EBV strains are less effective generators of LPD associated lesioning than the wild-type [2,11]. Lytic infection results in the secretion of paracrine factors that may stimulate the growth of latently infected B-cell lines [9, 11]. While it may have a more diffuse effect than the latent mechanism, EBV lytic infection mediated signalling may contribute to the development of lymphoproliferative disease.
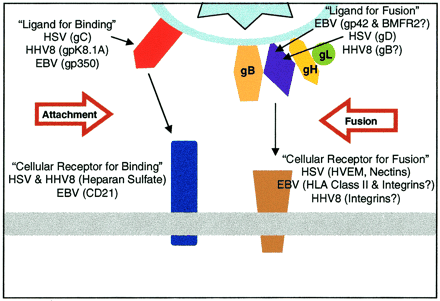
The lytic cycle begins with mature EBV virions reaching target host cells, such as B-cells. Contact between the virion and B-cell is initiated by the binding of EBV glycoprotein 350 to B-cell CD-21 [12]. For the most part, EBV infection is specific to cells expressing membranous CD-21 (namely B-cells and some epithelia), though some CD-21 independent attachment mechanisms are possible albeit with low efficacy [8]. For the purposes of this review, focus will be placed on viral infection of B-lymphocytes. EBV 350 is an example of a lytically expressed gene that is specifically target by the human immune system [13]. This gp350 binding is complemented by the binding of EBV gp42 to B-cell MHC-II [14]. In order for fusion to occur between the viral envelope and B-cell membrane, the EBV virion must have functional gH, gL, and gp42 spike glycoproteins (Figure 4)[15]. There is also evidence that a third glycoprotein, gB, contributes to membrane fusion [21].
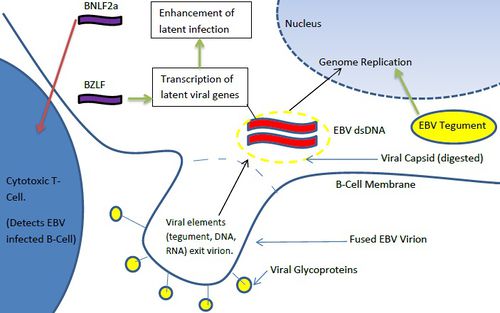
Once the gH/gL/gp42 complex penetrates the B-cell membrane, the viral capsid is dissolved, allowing the viral dsDNA genome to enter the cytoplasm. Interesting, viral RNA is often packaged in the virions in addition to the dsDNA genome. These transuced elements are referred to as tvRNA and include noncoding RNAs and mRNAs [16]. EBV tvRNA contribute to successful infection, cytokine signaling, and initiate transition towards a latent life cycle [16] (Figure 5). One of these mRNAs, a BZLF1 (a transcription factor) helps to activate viral promoters that trigger a transition to a pre-latent phase [16]. Another virion carried RNA, BNLF2a transcript, contributes to the protection of the infected B-cells from cytotoxic T-cells [16]. While cytotoxic T-cells are less adept at detecting BNLF2a(+) EBV infected cells [16], the exact mechanism by which this tvRNA reduces B-cell recognition is not known. In addition to tvRNA mediated B-cell alteration, viral tegument proteins also contribute to the progression of infection. EBV tegument released into the cytoplasm of host cells upregulates the transcription of IE (intermediate-early) genes which typically code for other transcription factors [9]. These factors subsequently upregulate genes involved in viral DNA replication [9]. Hence, virion transported viral RNA and tegument protein play immediate roles in altering B-cell character to ensure successful initial infection and immune system evasion. EBV tvRNA appear to directly contribute to transition from a lytic life cycle to a latent mode of proliferation.
In the lytic life cycle, viral dsDNA generally localizes to the nucleus for replication [17]. After viral dsDNA is replicated using host machinery, the production of viral structural proteins is upregulated, and viral particles are assembled in the nucleus [9,18]. After full particles are assembled in the nucleus, they bud out of the nuclear membrane, then the cell membrane. When exiting the host cell, these virions may acquire their primary envelope from the nuclear membrane, and the outer envelope from the cell membrane [19]. Unlike many other herpesviruses, this exit mechanism does not initiate mandatory cell death [19,20].
In conclusion, the lytic life cycle of the Epstein-Barr virus plays a less central role in the maintenance of viral infection than what is seen in closely related herpesviruses. However, the lytic life cycle is indispensable to host-host spread and horizontal intercellular transmission. In Epstein-Barr, lytic immune response avoidance mechanisms are less robust than those seen in other herpesviruses. This reflects the fact that EBV relies more on clonal replication of infected B-cells (latent mediated) for maintenance of an infection within a host. Virion carried proteins and RNA change the character of infected B-cells so as to maximize chances of successful infection, and in some cases to initiate latent mechanisms. Interestingly, lytic virion budding by EBV is not very lethal to host cells, a finding that has potential applications to putative anti-EBV agents.
Latent Life Cycle
While the lytic life cycle of EBV is vital to host-host transmission and had mechanisms that support successful infections within human hosts, the latent life cycle makes a more direct contribution to lymphoproliferative disease. In most cases, once EBV virions achieve primary infection of B-lymphocytes, the virus primarily replicates by a latent mechanism [2,16]. This results in the transformation of B-cells into ever-proliferating lymphoblastoid cell lines (LCL's) [2]. The latent phase in the EBV viral life cycle is defined by: 1)No production of virions, and 2) the production of a select few viral proteins and transcripts [22]. Many of these select latent viral proteins modulate the character of host B-cells and contribute to lymphoproliferation [2,22]. In contrast to the lytic cycle where the viral genome is brought to the nucleus and copied, under the lytic cycle infected B-cells contain several copies of extrachromosomal EBV episomes [2]. Hence, as the B-cells proliferate, so does the virus.
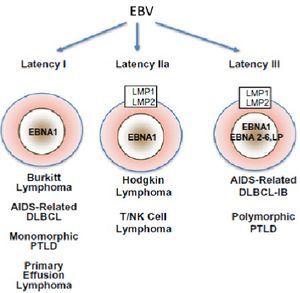
Latency can divided into 3 distinct stages defined by which latent genes are being expressed (Figure 6). In latency I, only EBNA1 is expressed [22], while in latency II EBNA1 is expressed along with intermediate levels of other EBNAs and LMP proteins [2,22]. Latency II can be subdivided into IIa, where EBNA2, but not LMP1 is expressed, and IIb, where the expression of these two proteins is reversed [23].Latency III is characterized by unregulated expression of the complete latent viral proteome which includes EBNAs 1-6, as well as 3 LMP proteins (1,2A, and 2B)[22]. The transcription of the circularized viral genome begins at either the Wp or Cp promoter (Figure 2)[2]. Differential splicing of the same long transcript (in green, figure 2) yields the different EBNA mRNA. In vivo LCLs typically display latency III-like expression profiles [22]. B-cells must have latency III expression profiles for successful generation of LCLs in vitro [23]. In addition to these viral proteins, several non-coding RNAs (EBERs) and micro-RNAs are also produced during all three stages of latency [2,7,22,24].
A broad and continually evolving area of research is centered on determining how these latent viral factors transform their B-cell hosts. Do different latent expression profiles result in different malignancies? What are the signaling cascades behind viral latent protein modulation of B-cell character? Why is the EBV latent mode of replication so successful in sustaining long-lasting infections in human hosts? Questions such as these have been well-studied, revealing an expansive and intricate mode of B-cell manipulation by EBV.
Latent Mechanisms Contributing to Immune Response Evasion
As noted above, once primary EBV infection establishes a small infected B-cell population in a host, further viral proliferation occurs primarily by way of a latent mechanism in which EBV is replicated in association with lymphoproliferation. Though the latent mechanism may afford a lower risk of detection by the host immune system than virion production, all EBV viral proteins are immunogenic [22]. As such, lymphoproliferative diseases associated with the high-expression type III latency occur primarily in immunocompromised hosts [22]. Type I and II latent infections may be successful in immunocompetent hosts due to lessened antigen presentation. In fact, EBNA1 may be capable of further reducing antigen presentation by way of inhibition of host HLA-1 activity (human MHC) [25]. Due to this combination of mechanisms, it appears that cytotoxic T-cells are generally incapable of EBNA1 recognition [26]. In addition, latent infection may induce the transcription of c-myc oncogene, resulting in a reduction of host immune-stimulating signaling [27]. Further, EBV may sustain successful infection even in the face of an extreme host immune response through adopting latency 0. During latency 0, the EBV genome is sequestered to the nucleus of infected cells and all expression is turned off, which essentially renders the virus undetectable [28]. This allows for the establishment of EBV reservoirs in the host B-cells that can survive an immune response firestorm.
Latent Membrane Protein 1 (LMP1)
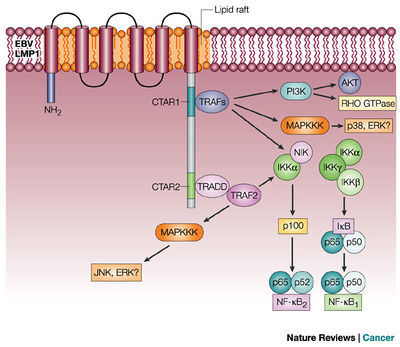
Latent Membrane Protein 1 is perhaps one of the most-well studied latent genes expressed by the Epstein-Barr Virus. This protein is a potent inducer of lymphoproliferative disease through a variety of cascades involving interactions with host cell proteins. LMP1 is expressed on the membrane of infected B-cells during latency type II and III [22]. Interestingly, LMP1's activity mimics that of a constitutively active tumor necrosis factor receptor, CD40 [22,2,29]. The lymphoproliferative capabilities of LMP1 are primarily centered around its ability to mimic the signaling profile of a constitutively active CD40. Hence, LMP-1 modulates several complex signaling cascades within host cells, many of which contribute to cell proliferation, growth, or suppression of apoptosis [2](Figure 6).
It is widely accepted that LMP-1 directly interacts with human Tumor Necrosis Factor Receptor associated proteins (TRAFs) [2]. These TRAFs bind the cytoplasmic carboxy terminus of LMP-1, which are termed C-terminal activation regions [2,30].The LMP-1 protein may be subdivided into 3 subdomains: membrane spanning, cytoplasmic N-Terminal, and cytoplasmic C-terminal, all of which are required for induction of lymphoproliferation in vivo [30](Figure 7).
EBNA1
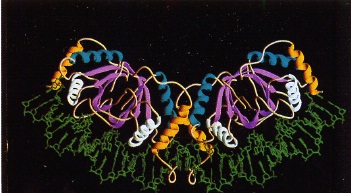
As noted above, EBNA1 is the only EBV viral protein expressed in all major types of latent infection [22,31]. EBNA1 is a viral nuclear antigen that binds specifically to the EBV episome in host cells. Interestingly, EBNA1 primarily exists in a dimeric form in vivo . EBNA1 has two primary domains: a DNA-specific domain that bind to the viral origin of replication (oriP), and a host-DNA tethering domain that associates with the human B-cell genome [32]. The crystal structure of EBNA1 has been resolved [33] and shows the presence of two unique binding domains (Figure 8). Interestingly, the N-terminal region of EBNA1 appears to wrap around a portion of the EBV DNA helix.
This binding between EBNA1 and the viral episome reflects a strong structure-function relationship. EBNA1 has been shown to act as an important "manager" of the EBV episome in latently infected cells. For example, EBNA1
References
[1]"Epstein-Barr Virus and Infectious Mononucleosis." Centers for Disease Control. http://www.cdc.gov/ncidod/diseases/ebv.htm. Accessed: 11/3/12.
[2]Young LS,Rickinson AB. 2004. Epstein-Barr Virus: 40 Years On. Nature Reviews. 4:757-768.
[3]Wang D, Liebowitz D, Kieff E. 1985. An EBV Membrane Protein Expressed in Immortalized Lymphocytes Transforms Established Rodent Cells. Cell. 43: 831-840.
[4]Epstein MA, Achong BG, Barr YM. 1964. Virus Particles In Cultured Lymphoblasts from Burkitt's Lymphoma. The Lancet. 283:702-703.
[5] Baer R, Bankier AT, Biggin MD, Deininger DL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, Tuffnell PS, Barrell BG. 1984. DNA sequence and expression of the B95-8 Epstein-Barr Virus Genome. Nature. 310: 207-211.
[6] Mahiet C, Ergani A, Huot N, Alende N, Azough A, Salvaire F, Bensimon A, Conseiller E, Wain-Hobson S, Labetoulle M, Berradeau S. 2012. Structural Variability of the Herpes Simplex Virus 1 Genome in Vitro and In Vivo. Journal of Virology. 86(16): 8592-8601.
[7] "Epstein-Barr Virus." International Agency for Research on Cancer. IARC Monographs. 1997. 49-92. http://monographs.iarc.fr/ENG/Monographs/vol100B/mono100B-6.pdf. Accessed: 11/3/12.
[8]Liebowitz D, Kieff E. 1993. Epstein-Barr virus. In: The Human Herpesvirus. Roizman B, Whitley RJ, Lopez C, editors. New York. 107–172.
[9] Swaminathan S, Kenney S. "The Epstein-Barr Virus Lytic Life Cycle." 2009. In: DNA Tumor Viruses. 285-315.
[10] Steven, N. M., Annels, N. E., Kumar, A., Leese, A. M., Kurilla, M. G. and Rickinson, A. B. 1997. Immediate early and early lytic cycle proteins are frequent targets of the EpsteinBarr virus-induced cytotoxic T cell response. J Exp Med. 185: 1605–1617.
[11] Hong, G. K., Gulley, M. L., Feng, W. H., Delecluse, H. J., Holley-Guthrie, E. and Kenney, S.C. 2005. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. Journal of Virology. 79: 13993–14003.
[12] Nemerow, G. R., Mold, C., Schwend, V. K., Tollefson, V. Cooper, N. R. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein–Barr virus(EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complment fragment C3d. J. Virol. 61: 1416–1420.
[13] Khyatti M, Patel PC, Stefanescu I, Menezes J. 1991. Epstein-Barr virus (EBV) glycoprotein gp350 expressed on transfected cells resistant to natural killer cell activity serves as a target antigen for EBV-specific antibody-dependent cellular cytotoxicity. Journal of Virology. 65(2): 996-1001.
[14] Borza, C. M. Hutt-Fletcher, L. M. 2002. Alternate replication in B-cells and epithelial cells switches tropism of Epstein–Barr virus. Nature Med. 8: 594–599.
[15] Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. 2006. Soluble Epstein-Barr Virus Glycoproteins gH, gL, and gp42 Form a 1:1:1 Stable Complex That Acts Like Soluble gp42 in B-Cell Fusion but Not in Epithelial Cell Fusion. Journal of Virology. 80(19): 9444-9454.
[16]Jochum S, Ruiss R, Moosman A, Hammershmidt W, Zeidler R. 2012. RNAs in Epstein-Barr virions control early steps of infection. PNAS. 109(21): 7970-7971.
[17] Daikoku T, Kudoh A, Fujita M, et al. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. Journal of Virology.79(6):3409–3418.
[18] Henson BW, Perkins EM, Cothran JE, Desai P. 2009. Self-assembly of Epstein-Barr virus capsids. J Virol. 83:3877–90.
[19]Gong M, Kieff E. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. 64(4):1507-1516.
[20] "Epstein-Barr Virus." http://virology-online.com/viruses/EBV.htm Accessed: 11/10/12.
[21] Spear PG, Longnecker R. 2003. Herpesvirus Entry: An Update. Journal of Virology. 77(19): 10179.
[22] Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients.Cancer Letters. 305: 163-174.
[23] E Klein, LL Kis, G Klein. 2007. Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions.Oncogene. 26:1297–1305.
[24] Fok V, Friend K, Seitz JA. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. Journal of Cell Biology. 173(3): 319-325.
[25] Kitagawa N, Goto M, Kurozumi K, Maruo S, Fukayama M, Naoe T, Yasukawa M, Hino K, Suzuki T, Todo S, Takada K. 2000. Epstein-Barr virus-encoded poly(A)(-) RNA supports Burkitt’s lymphoma growth through interleukin-10 induction. Embo J. 19: 6742–6750.
[26] Hochberg D, Middeldorp JM, Catalina M, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. PNAS. 101(1): 239-244.
[27] God JM, Haque A. 2010. Burkitt lymphoma: pathogenesis and immune evasion, J. Oncol.
[28]Ning S. 2001. Innate immune modulation in EBV infection. Herpesviridae. 2:1.
[29] Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. 1997.Latent membrane protein 1 of Epstein–Barr virus mimics a constitutively active receptor molecule. The EMBO Journal. 16(20): 6131–6140.
[30] Brodeur SR, Cheng G, Baltimore D, Thorley-Lawson DA. 1997. Localization of the Major NF-kB-activating Site and the Sole TRAF3 Binding Site of LMP-1 Defines Two Distinct Signaling Motifs. Journal of Biological Chemistry. 272(32): 19777–19784.
[31]Gruhne B, Sompalle R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. 2009. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. PNAS. 106(7): 2313-2318.
[32] Saha A, Robertson E. 2011. Epstein-Barr Virus–Associated B-cell Lymphomas: Pathogenesis and Clinical Outcomes. Clinical Cancer Research. 17(10): 3056-3063.
[33] Bochkarev A, Barwell JA, Pueftzner RA, Furey Jr. W, Edwards AM, Frappier L. 1995. Crystal Structure of the DNA-Binding Domain of the Epstein-Barr Virus Origin-Binding Protein EBNA1. Cell. 83: 39-46.
