Trichophyton rubrum: Difference between revisions
| (60 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
===Species=== | ===Species=== | ||
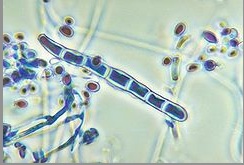
''Genus: Trichophyton; Species: T. rubrum'' | [[Image:Trichophytonjpegg.jpg|frame|right|150px|Microscopic view of microconidia and macroconidia in ''T. rubrum''. Credit: CDC/Dr. Libero Ajello]] | ||
Genus: ''Trichophyton''; Species: ''T. rubrum'' | |||
==Description and significance== | ==Description and significance== | ||
''Trichophyton rubrum'', is the most common causitive agent of dermatophytosis worldwide, mainly occupying the humans’ feet, skin, and between fingernails [3]. ''T. rubrum'' is known to be one of the most prominent anthrophilic species of dermatophtyes [1], a fungus commonly causing skin diseases, appearing in various shades of white, yellow, brown, and red. It may also be found in various textures, being waxy, cottony, or smooth. Even though it is commonly observed, ''T. rubrum'' infections are incredibly hard to diagnose, and difficult to differentiate from other dermatophytes. Because this fungal pathogen is poorly understood, the discovery of its genomic structure may significantly reduce the health costs of those who suffer various forms of dermatophytosis caused by ''T. rubrum''. Through validated information of what is known on its genome sequence, ''T. rubrum'' is a keratinophilic filamentous fungus. | |||
==Genome structure== | ==Genome structure== | ||
In the absence of complete m-RNA-based evidence, the complexity of filamentous fungi gene structures make gene interpretations challenging [1]. Due to the lack of biochemical identification techniques available, pleomorphism, and cultural variability of Dermatophytes, the current knowledge of the ''T. rubrum'' genomic sequencing is limited but is in progress. | |||
Thus far, the cloning of cDNAs have enabled the generation of expressed sequence tags (ESTs) and have been found to effectively attribute to the identification processes undertaken [4]. The ''T. rubrum'' genome has now been organized into five chromosomes. Altogether, estimated to be 22Mb in which 5 to 10% of the genome are repetitive DNA subunits of 8 and 50% AT content [4]. To date, 43 unique nuclear-encoded genes have been analyzed, and approximately more than half are proteases [3]. | |||
==Cell and colony structure== | |||
Due to pleomorphism, many strains and varieties of ''T. rubrum'' have been described. ''T. rubrum'' colonies typically produce a white to cream color pigmentation on its surface and have a reverse side that ranges from yellow-brown to wine-red [7]. Textures vary among the diverse strains of ''T. rubrum'' in which colony structures can appear flat to slightly raised [7]. Most cultures have been identified to be granular strains, grain-shaped [7]. Granular strains include abundant numbers of clavate (oblong with thick ends) to pyriform (pear-shaped) microconidia, small spores produced via asexual reproduction. On the contrary, the production of macroconidia (large spores produced via asexual reproduction) are scanty to moderate, have thin walled septa, and are bacillus-shaped [7]. | |||
==Metabolism== | |||
''T. rubrum's'' metabolism is highly influenced and responsive to environmental pH conditions that is sensed. Based on recent studies and annotations, more than half of the ''T. rubrum'' genome sequence is composed of proteases, mostly keritinases[3]. The production of these proteases depend on the pH content of keratins secreted [6] and is optimal in acidic conditions [4], thus determining ''T. rubrum's'' virulence factors (i.e. ability to ingest keratin for growth and strength) [3]. | |||
==Ecology== | |||
''T. rubrum'' is an anthrophilic dermatophyte, known to inhabit moist areas of the human skin, where skin folds, or even nails, where keratin is abundant for its growth and survival [1]. ''T. rubrum'' may also contaminate items such as clothing or bedding. Though much about ''T. rubrum'' is still unknown, through the identification processes of its genome sequence that have been undertaken it appears that some of its gene expression had been linked to the presence of cytotoxic drugs [5]. For the time being, ''T. rubrum'' may not be making any direct contributions to the environment, but the studies of it may lead to many technological advances and methodologies to diagnose other forms of dermatophytosis [1]. | |||
==Pathology== | |||
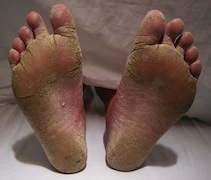
[[Image:FeetFungal.JPG|frame|right|150px| Severe case of athlete's foot. Credit: James Heilman, MD]] | |||
''T. rubrum'' is the most common infectious, anthrophilic species of dermatophytes [1]. Very little is known about the mechanism of its invasion and pathogenicity [5]. Though it is usually not life-threatening, infections are long-lasting, recurring, and incredibly difficult to cure. The fungal pathogen’s ability to produce and secrete proteolytic enzymes is a major virulence factor [6]. ''T. rubrum'' attacks the human skin and nails via keratin degradation [1]. Keratin, a fibrous protein, is a major structural component of the human skin and nails; thus ''T. rubrum'' invades through the stratum corneum, the outermost layer of the epidermis, to obtain keratin. This invasion causes dermatophytosis infections such as athlete’s foot, fungal infections between fingernails, jock itch, and may be painful. The infections may be transmitted from person to person. Anti-fungal medications are available to prevent infections, however infections typically reoccur. | |||
== | ==References== | ||
T. | 1. White, T., Henn, M., et al., Genomic Determinants of Infection in Dermatophyte Fungi. The Fungal Genome Initiative, Mar. 2012 | ||
http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/Dermatophyte_WP_seq.pdf | |||
2. De Biervre, C. and Dujon, B. Organisation of the mitochondrial genome of ''Trichophyton rubrum''. Current Genetics, Volume 26, Issue 6: 553-559. | |||
3. Yang, J., Chen, L., Wang, L., et al., TrED: the ''Trichophyton rubrum'' Expression Database. BMC Genomics, Volume 8: 250. | |||
4. Silveira, H.C.S., Gras D.E., Cazzaniga, R.A., et al. “Transcriptional profiling reveals genes in the human pathogen...”. Microbial Pathogenesis, Volume 48, Issue 2, 2010: 91-96. | |||
5. Maranhao, F.C.A., Paniao, F.G., Martinez-Rossi, N.M.. “Isolation of transcripts over-expressed in human pathogen...”. Microbial Pathogenesis, Volume 43, Issue 4, 2007: 166- 172. | |||
6. Chen, J., Yi, Jinling, Liu, Li, et al. “Substrate adaptation of ''Trichophyton rubrum''...”. Microbial Pathogenesis, Volume 48, Issue 2, 2010: 57-61. | |||
7. Ellis, D., ''Trichophyton rubrum'' granular strain. Mycology Online, Apr. 2012 | |||
http://www.mycology.adelaide.edu.au/Fungal_Descriptions/Dermatophytes/Trichophyton/rubrum.html | |||
Edited by Melinda Dao of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio | Edited by Melinda Dao of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio | ||
Latest revision as of 03:51, 7 May 2012
A Microbial Biorealm page on the genus Trichophyton rubrum
Classification
Higher order taxa
Domain: Eukaryota; Kingdom: Fungi; Phylum: Ascomycota; Class: Eurotiomycetes; Order: Onygenales; Family: Arthrodermactaceae
Species
Genus: Trichophyton; Species: T. rubrum
Description and significance
Trichophyton rubrum, is the most common causitive agent of dermatophytosis worldwide, mainly occupying the humans’ feet, skin, and between fingernails [3]. T. rubrum is known to be one of the most prominent anthrophilic species of dermatophtyes [1], a fungus commonly causing skin diseases, appearing in various shades of white, yellow, brown, and red. It may also be found in various textures, being waxy, cottony, or smooth. Even though it is commonly observed, T. rubrum infections are incredibly hard to diagnose, and difficult to differentiate from other dermatophytes. Because this fungal pathogen is poorly understood, the discovery of its genomic structure may significantly reduce the health costs of those who suffer various forms of dermatophytosis caused by T. rubrum. Through validated information of what is known on its genome sequence, T. rubrum is a keratinophilic filamentous fungus.
Genome structure
In the absence of complete m-RNA-based evidence, the complexity of filamentous fungi gene structures make gene interpretations challenging [1]. Due to the lack of biochemical identification techniques available, pleomorphism, and cultural variability of Dermatophytes, the current knowledge of the T. rubrum genomic sequencing is limited but is in progress. Thus far, the cloning of cDNAs have enabled the generation of expressed sequence tags (ESTs) and have been found to effectively attribute to the identification processes undertaken [4]. The T. rubrum genome has now been organized into five chromosomes. Altogether, estimated to be 22Mb in which 5 to 10% of the genome are repetitive DNA subunits of 8 and 50% AT content [4]. To date, 43 unique nuclear-encoded genes have been analyzed, and approximately more than half are proteases [3].
Cell and colony structure
Due to pleomorphism, many strains and varieties of T. rubrum have been described. T. rubrum colonies typically produce a white to cream color pigmentation on its surface and have a reverse side that ranges from yellow-brown to wine-red [7]. Textures vary among the diverse strains of T. rubrum in which colony structures can appear flat to slightly raised [7]. Most cultures have been identified to be granular strains, grain-shaped [7]. Granular strains include abundant numbers of clavate (oblong with thick ends) to pyriform (pear-shaped) microconidia, small spores produced via asexual reproduction. On the contrary, the production of macroconidia (large spores produced via asexual reproduction) are scanty to moderate, have thin walled septa, and are bacillus-shaped [7].
Metabolism
T. rubrum's metabolism is highly influenced and responsive to environmental pH conditions that is sensed. Based on recent studies and annotations, more than half of the T. rubrum genome sequence is composed of proteases, mostly keritinases[3]. The production of these proteases depend on the pH content of keratins secreted [6] and is optimal in acidic conditions [4], thus determining T. rubrum's virulence factors (i.e. ability to ingest keratin for growth and strength) [3].
Ecology
T. rubrum is an anthrophilic dermatophyte, known to inhabit moist areas of the human skin, where skin folds, or even nails, where keratin is abundant for its growth and survival [1]. T. rubrum may also contaminate items such as clothing or bedding. Though much about T. rubrum is still unknown, through the identification processes of its genome sequence that have been undertaken it appears that some of its gene expression had been linked to the presence of cytotoxic drugs [5]. For the time being, T. rubrum may not be making any direct contributions to the environment, but the studies of it may lead to many technological advances and methodologies to diagnose other forms of dermatophytosis [1].
Pathology
T. rubrum is the most common infectious, anthrophilic species of dermatophytes [1]. Very little is known about the mechanism of its invasion and pathogenicity [5]. Though it is usually not life-threatening, infections are long-lasting, recurring, and incredibly difficult to cure. The fungal pathogen’s ability to produce and secrete proteolytic enzymes is a major virulence factor [6]. T. rubrum attacks the human skin and nails via keratin degradation [1]. Keratin, a fibrous protein, is a major structural component of the human skin and nails; thus T. rubrum invades through the stratum corneum, the outermost layer of the epidermis, to obtain keratin. This invasion causes dermatophytosis infections such as athlete’s foot, fungal infections between fingernails, jock itch, and may be painful. The infections may be transmitted from person to person. Anti-fungal medications are available to prevent infections, however infections typically reoccur.
References
1. White, T., Henn, M., et al., Genomic Determinants of Infection in Dermatophyte Fungi. The Fungal Genome Initiative, Mar. 2012 http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/Dermatophyte_WP_seq.pdf
2. De Biervre, C. and Dujon, B. Organisation of the mitochondrial genome of Trichophyton rubrum. Current Genetics, Volume 26, Issue 6: 553-559.
3. Yang, J., Chen, L., Wang, L., et al., TrED: the Trichophyton rubrum Expression Database. BMC Genomics, Volume 8: 250.
4. Silveira, H.C.S., Gras D.E., Cazzaniga, R.A., et al. “Transcriptional profiling reveals genes in the human pathogen...”. Microbial Pathogenesis, Volume 48, Issue 2, 2010: 91-96.
5. Maranhao, F.C.A., Paniao, F.G., Martinez-Rossi, N.M.. “Isolation of transcripts over-expressed in human pathogen...”. Microbial Pathogenesis, Volume 43, Issue 4, 2007: 166- 172.
6. Chen, J., Yi, Jinling, Liu, Li, et al. “Substrate adaptation of Trichophyton rubrum...”. Microbial Pathogenesis, Volume 48, Issue 2, 2010: 57-61.
7. Ellis, D., Trichophyton rubrum granular strain. Mycology Online, Apr. 2012 http://www.mycology.adelaide.edu.au/Fungal_Descriptions/Dermatophytes/Trichophyton/rubrum.html
Edited by Melinda Dao of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio