User:S4355889
Name: Callum Le Lay
Bench ID: C
Date: 31/08/2016
[1]
Classification
Higher order taxa
Bacteria - Terrabacteria group - Firmicutes - Negativicutes - Veillonellales - Veillonellaceae - Veillonella
Species
Type strain: Prevot Te 3 = ATCC 10790 = DSM 2008 = JCM 12972
Description and significance
Named after the french biologist Adrien Veillon, who first discovered the species in 1898[2][3], Veillonella parvula is a gram negative bacteria found in the human oral cavity[4][5] and the gastrointestinal tract[6]. The Veillonella genus is part of the Negativicutes class of bacteria which are known for their peculiar gram negative cell wall, which they have despite being a part of the Firmicutes phylum in which the majority of species are gram positive. V. parvula is obligately anaerobic, auxotrophic, lactate fermenting and cocci shaped [7][8][9][4]. The species is small at 0.3-0.5µm[7].
Veillonella make close physical associations with species of Streptococcus genus[10]. Veillonella cannot metabolise carbohydrates[11] and because of this will ferment other sources of energy. V. parvula ferments lactate, a common byproduct of anaerobic respiration in bacteria. It will bind to the surface of other fermenting bacteria and metabolise the lactate as it is produced[12][10]. This benefits V. parvula as it does not have to compete for resources. Coaggregation of Veillonella species with certain Streptococcus species (each species has preferences) is also shown to promote biofilm formation[10] and the two species are known early colonisers in oral plaque communities[12].
The importance of understanding this species of bacteria is its role in oral plaques. It has been shown to produce biofilm[10] and plays a role indetermining the composition of the plaque community. Also it V. parvula can be opportunistically pathogenic, meaning it will cause disease when the hosts immune system is compromised. It can be present in dental caries[REF] and in chronic peridontitis[13][14][13]; and is less commonly associated with other types of disease such as endodontitus[REF], vaginosis[15] and even osteomyetitis[16]. FIX LAST PARAGRAPH
Genome structure
Veillonella parvula, specifically the type strian DSM 2008, was sequenced and analysed in 2009 by Gronow et. al[7]. The genome is circular and is 2.13Mbp long. It contains 1920 genes, 97% being protein coding genes and 3 % being RNA coding genes. This means the density of protein coding genes is about one gene per 1.1kbp. 0.78% of the protein coding genes were predicted to be pseudogenes. The GC content was found to be 39% and there were no detected extrachromosomal elements.
Comparing to average bacterial properties[17], the genome size of V. parvula is normal but its protein coding density is quite low. This is not uncommon for members of the Firmicutes phylum[18][19].
Cell structure and metabolism
Vellonellaceae do not gram stain positive like other species of the Firmicutes phylum. They stain gram negative[3][7][21][9] and have the cell wall composition common of gram negatives: a lipopolysaccharide coated outer membrane, a thin peptidoglycan layer and an inner cytoplasmic membrane[22][23]. Vellonella have a characteristic diamine containing peptidoglycan layer. Two diamine compounds are required for cell growth because of this: cardaverine and putrescine. These two molecules are required for maintenance of the diamine containing peptidoglycan and the integrity of the cell envelope[24]. Veillonella also contain plamalogens in their cytoplasmic membrane, which are molecules used to regulate fluidity. Plasmenylethanolamine and plasmenylserine are plasmalogens present in V. parvula [21].
Hexokinase is required for the first step in glycolysis. Veillonella have no detectable hexokinase activity [11], which partially explains why the genus is unable to metabolise glucose. Research by Rogosa et. al. shows that the rest of the glycolytic system in Veillonella is functional. The researchers suggest that the pathway could instead be a viable route for the production of glucose-6-phosphate through gluconeogenesis, because most of the pathway is functional.
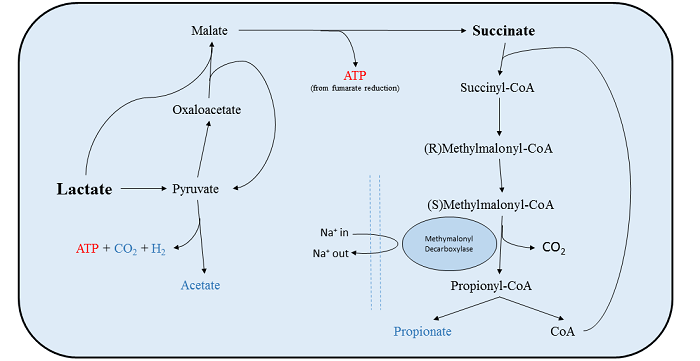
V. parvula uses lactate, malate and formate as a source of energy, which it metabolises anaerobically to produce acetate, carbon dioxide, hydrogen gas and propionate. It metabolises these compounds by converting them to pyruvate before degrading them to acetate, producing about 1 ATP molecule per mole of lactate[25][20][22]. In addition to this, V. parvula is able to take up succinate as a co-factor (and only as a cofactor) to improve growth rate. In V. parvula about 50% of lactate that is metabolised is oxidised to pyruvate then degraded to actetate. The other half is converted to succinate (malate and formate are intermediates) and then degraded to propionate by methlmalonyldecarboxylase (MMD). This reaction does not yield a net increase in ATP, but the reaction that the membrane bound MMD catalyzes is coupled to the production of a sodium electrochemical gradient[20]. In this way the bacteria is able to conserve the energy normally used to produce this gradient, meaning growth rates are increased when succinate is taken up in conjunction with lactate/malate/formate[25].
Ecology
V. parvula is a mesophilic, heterotrophic and obligately anaerobic organism[3][7]. It is found in humans and is most abundant within the intestinal tract[6] and within the oral cavity [4][5]. Veillonella species have been found within the vaginal community[15], but V. parvula has not been specifically identified as present. Upon infection, V. parvula can relocate to the bone/bone marrow[16] or other environments where it is not commonly found in.
There does not seem to be much interaction between the host and V. parvula, as it is commensal, instead the bacteria interacts heavily with other bacterial species that in turn may interact with the host [12].
Veillonella can utilise the lactate in the saliva of the oral cavity, but they have a higher grhttps://microbewiki.kenyon.edu/index.php/User_talk:S4355889owth rate when they are partnered to a lactate producing bacteria. They can form partnerships with species such as Actinomyces oris, Porphyromonas gingervalis [perisamy->21] and any of the Streptococcus species[12], in which both partners have a higher growth rate than they would alone. The species that Veillonella bind with affects whether there is an increase in growth rate, and the extent of that increase, for the two species. Perisamy[12] examined the relationships that Veillonella had with other species and found that the effect on growth caused by two-way species interactions did not predict the effect in three-way interactions. For example: though the researchers found that A. oris, P. gingervalis and Veillonella species could all interact for greater growth in two-way interactions, there was no increase in a community composed of all three species. This shows that Veillonella have high specificity in regards to which species they will interact with for efficient utilisation of resources. It is because of this specificity and its position as an early plaque coloniser, that Veillonella plays a very important role in the plaque community. It dictates which of the later colonising species will have a greater abundance in the plaque.
V. parvula induces greater growth and biofilm production in the Streptococcus species mutans and sanguinus[10]. It has been associated with both healthy[14] and unhealthy plaque communities[13].
Pathology
Veillonella have a commensal relationship with humans, but can, in rare cases, be opportunistically pathogenic. In these cases, the species identified is commonly V. parvula. V. parvula has been found to cause osteomyelitis[16], sepsis and bacteraemia[26][27][28], spondylodiscitis[29][30], urinary tract infection[31] and endocarditis[32]. Instances of V. parvula associated disease, as listed, are very rare in healthy patients[33]. The host in almost every case has had a compromised immune system, commonly through diabetes, HIV, old age, pregnancy or some recessive disorder. It should be noted that it is rare to find instances of disease where Veillonella are a major contributor, but many polymicrobial infections are likely to have Veillonella present, given its importance in biofilm generation and multispecies interaction.
Application to biotechnology
As V. parvula is not usually pathogenic there is not much funded research being done to find drug targets for it. As a model organism, V. parvula is good: it is often found in high abundance in the oral cavity and it is the best understood of the Veillonella, but it is not a great model for gram negative bacteria and there are already better models (Pseudomonas aeruginosa) available for biofilm formation.
There have been some attempts to develop technologies that would help researchers better understand the biology of the Veillonella genus. A study by Liu et. al.[34] looks at developing transformation techniques in V. parvula with the aim of better understanding Veillonellaceae biology. A paper from 1975 showed that Veillonella can be made to fluoresce under long wave ultraviolet light[35], which is useful for segregating species of the genus from a community.
Current research
Summarise some of the most recent discoveries regarding this species.
References
- ↑ MICR3004
- ↑ List of prokaryotic names with standing in nomenclature
- ↑ 3.0 3.1 3.2 Veillon A, Zuber MM. Recherches sur quelques microbes strictement anaérobies et leur rôle en pathologie. Arch Med Exp 1898; 10:517-545.
- ↑ 4.0 4.1 4.2 Edlund, A., Yang, Y., Yooseph, S., Hall, A. P., Nguyen, D. D., Dorrestein, P. C., . . . McLean, J. S. (2015). Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J, 9(12), 2605-2619. doi:10.1038/ismej.2015.72
- ↑ 5.0 5.1 Mashima, I., Kamaguchi, A., & Nakazawa, F. (2011). The distribution and frequency of oral veillonella spp. in the tongue biofilm of healthy young adults. Curr Microbiol, 63(5), 403-407. doi:10.1007/s00284-011-9993-2
- ↑ 6.0 6.1 van den Bogert, B., Erkus, O., Boekhorst, J., de Goffau, M., Smid, E. J., Zoetendal, E. G., & Kleerebezem, M. (2013). Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol, 85(2), 376-388. doi:10.1111/1574-6941.12127
- ↑ 7.0 7.1 7.2 7.3 7.4 Gronow, S., Welnitz, S., Lapidus, A., Nolan, M., Ivanova, N., Glavina Del Rio, T., . . . Lucas, S. (2010). Complete genome sequence of Veillonella parvula type strain (Te3). Stand Genomic Sci, 2(1), 57-65. doi:10.4056/sigs.521107
- ↑ http://www.genome.jp/dbget-bin/www_bget?vpr:Vpar_1247+vpr:Vpar_1248+vpr:Vpar_1762+vpr:Vpar_1763
- ↑ 9.0 9.1 9.2 Delwiche, E. A., Pestka, J. J., & Tortorello, M. L. (1985). The Veillonellae: Gram-Negative Cocci with a Unique Physiology. Annual Reviews, 39, 18.
- ↑ 10.0 10.1 10.2 10.3 10.4 Mashima, I., & Nakazawa, F. (2014). The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe, 28, 54-61. doi:10.1016/j.anaerobe.2014.05.003
- ↑ 11.0 11.1 Rogosa, M., Krichevsky, M. I., & Bishop, F. S. (1965). Truncated Glycolytic System in Veillonella. Journal of Bacteriology, 90(1), 7.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 Periasamy, S., & Kolenbrander, P. E. (2010). Central Role of the Early Colonizer Veillonella sp. in Establishing Multispecies Biofilm Communities with Initial, Middle, and Late Colonizers of Enamel. Journal of Bacteriology, 192(12), 2965-2972. doi:10.1128/jb.01631-09
- ↑ 13.0 13.1 13.2 Silva-Boghossian, C. M., Neves, A. B., Resende, F. A., & Colombo, A. P. (2013). Suppuration-associated bacteria in patients with chronic and aggressive periodontitis. J Periodontol, 84(9), e9-e16. doi:10.1902/jop.2013.120639
- ↑ 14.0 14.1 Stingu, C. S., Jentsch, H., Eick, S., Schaumann, R., Knofler, G., & Rodloff, A. (2012). Microbial profile of patients with periodontitis compared with healthy subjects. Quintessence International, 43(2), 9.
- ↑ 15.0 15.1 Africa, C. W., Nel, J., & Stemmet, M. (2014). Anaerobes and bacterial vaginosis in pregnancy: virulence factors contributing to vaginal colonisation. Int J Environ Res Public Health, 11(7), 6979-7000. doi:10.3390/ijerph110706979
- ↑ 16.0 16.1 16.2 Al-Otaibi, F. E., & Al-Mohizea, M. M. (2014). Non-vertebral Veillonella species septicemia and osteomyelitis in a patient with diabetes: a case report and review of the literature. Journal of Medical Case Reports, 8(365), 5. doi:10.1186/1752-1947-8-365
- ↑ Koonin, E. V., & Wolf, Y. I. (2008). Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Research, 36(21), 6688-6719. doi:10.1093/nar/gkn668
- ↑ Schofeild, B. J., Skarshewski, A., Lachner, N., Ouwerkerk, D., Klieve, A. V., Dart, P., & Hugenholtz, P. (2016). Near complete genome sequence of the feed probiotic, Bacillus amyloliquefaciens H57. Standards in Genomic Sciences, 1(89), 7.
- ↑ Riedel, T., Bunk, B., Wittmann, J., Thürmer, A., Spröer, C., Gronow, S., . . . Overmann, J. (2015). Complete Genome Sequence of theClostridium difficileType Strain DSM 1296T. Genome Announcements, 3(5), e01186-01115. doi:10.1128/genomeA.01186-15
- ↑ 20.0 20.1 20.2 Dimroth, P. (1987). Sodium Ion Transport Decarboxylases and Other Aspects of Sodium Ion Cycling in Bacteria. Microbiological Reviews, 51(3), 20.
- ↑ 21.0 21.1 Olsen, I. (1997). Salient structural features in the chemical composition of oral anaerobes, with particular emphasis on plasmalogens and sphingolipids. Reviews in Medical Microbiology, 8(1), 3.
- ↑ 22.0 22.1 Delwiche, E. A., Pestka, J. J., & Tortorello, M. L. (1985). The Veillonellae: Gram-Negative Cocci with a Unique Physiology. Annual Reviews, 39, 18.
- ↑ Jumas-Bilak, E., Carlier, J. P., Jean-Pierre, H., Teyssier, C., Gay, B., Campos, J., & Marchandin, H. (2004). Veillonella montpellierensis sp. nov., a novel, anaerobic, Gram-negative coccus isolated from human clinical samples. Int J Syst Evol Microbiol, 54(Pt 4), 1311-1316. doi:10.1099/ijs.0.02952-0
- ↑ Kamio, Y., & Nakamura, K. (1987). Putrescine and Cadaverine Are Constituents of Peptidoglycan in Veillonella alcalescens and Veillonella parvula. Journal of Bacteriology, 169(6), 3.
- ↑ 25.0 25.1 Denger, K., & Schink, B. (1992). Energy conservation by succinate decarboxylation in Veillonella parvula. Journal of General Microbiology, 138(5), 4.
- ↑ Yagihashi, Y., & Arakaki, Y. (2012). Acute pyelonephritis and secondary bacteraemia caused by Veillonella during pregnancy. BMJ Case Rep, 2012. doi:10.1136/bcr-2012-007364
- ↑ Strach, M., Siedlar, M., Kowalczyk, D., Zembala, M., & Grodzicki, T. (2006). Sepsis caused by Veillonella parvula infection in a 17-year-old patient with X-linked agammaglobulinemia (Bruton's disease). J Clin Microbiol, 44(7), 2655-2656. doi:10.1128/JCM.00467-06
- ↑ Arrosagary, P. M., Salas, C., Morales, M., Correas, M., Barros, J. M., & Cordon, M. L. (1987). Bilateral Abscessed Orchiepididymitis Associated with Sepsis Caused by Veillonella parvula and Clostridium perfringens: Case Report and Review of the Literature. Journal of Clinical Microbiology, 25(8), 2.
- ↑ Kishen, T. J., Lindstrom, S. T., Etherington, G., & Diwan, A. D. (2012). Veillonella spondylodiscitis in a healthy 76-year-old lady. Eur Spine J, 21 Suppl 4, 413-417. doi:10.1007/s00586-011-1871-x
- ↑ Marriott, D., Stark, D., & Harkness, J. (2007). Veillonella parvula discitis and secondary bacteremia: a rare infection complicating endoscopy and colonoscopy? J Clin Microbiol, 45(2), 672-674. doi:10.1128/JCM.01633-06
- ↑ Berenger, B. M., Chui, L., Borkent, A., & Lee, M. C. (2015). Anaerobic urinary tract infection caused by Veillonella parvula identified using cystine-lactose-electrolyte deficient media and matrix-assisted laser desorption ionization-time of flight mass spectrometry. IDCases, 2(2), 44-46. doi:10.1016/j.idcr.2015.02.002
- ↑ Oh, S., Havlen, P. R., & Hussain, N. (2005). A Case of Polymicrobial Endocarditis due to Anaerobic Organisms in an Injection Drug User. Journal of General Internal Medicine, 20(10), 958-958. doi:10.1111/j.1525-1497.2005.0176.x
- ↑ Brook, I. (1996). Veillonella Infections in Children. Journal of Clinical Microbiology, 34(5), 2.
- ↑ Liu, J., Merritt, J., & Qi, F. (2011). Genetic transformation of Veillonella parvula. FEMS Microbiol Lett, 322(2), 138-144. doi:10.1111/j.1574-6968.2011.02344.x
- ↑ Chow, A. W., Patten, V., & Guze, L. B. (1975). Rapid Screening of Veillonella by Ultraviolet Fluorescence. Journal of Clinical Microbiology, 2(6), 3.
Cite error: <ref> tag defined in <references> has no name attribute.
Notes
TEMPORARY: TO BE DELETED AFTER FINISH
- https://www.mediawiki.org/wiki/Help:Formatting
- What type of referencing is wanted?
- What is the weird micr3004 reference?
HILPERT, W. & DIMROTH, P. (1982). Conversion of the chemical energy of methylmalonyl-CoA decarboxylation into a Na+ gradient. Nature, London 2%, 584-585. HILPERT, W. & DIMROTH, P. (1991). On the mechanism of sodium ion translocation by methylmalonyl-CoA decarboxylasef from Veillonella alcalescens. European Journal of Biochemistry 195, 79-86.
This page was written by Callum Le Lay for the MICR3004 course, Semester 2, 2016

