User:S4414061: Difference between revisions
| (46 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
===Species=== | ===Species=== | ||
'''Kingella oralis''' | <i>'''Kingella oralis'''</i> | ||
Type strain: strain UB-38 = ATCC 51147= CCUG 30450 = CIP 103803 | Type strain: strain UB-38 = ATCC 51147= CCUG 30450 = CIP 103803 | ||
==Description and significance== | ==Description and significance== | ||
Kingella oralis was first obtained from human periodontal isolates recovered from a purported <i>Eikenlla corrodens</i>-selective medium<sup>1</sup>, | <i>Kingella oralis</i>, <i>K. oralis</i> was first obtained from human periodontal isolates recovered from a purported <i>Eikenlla corrodens</i>-selective medium<sup>[[#References|1]]</sup>. In 1990, it was originally described as <i> Eikenella corrodens</i>-like isolates<sup>[[#References|2]]</sup>. Using 16sRNA sequencing, these isolates were identified as <i>K. orale</i> in 1993<sup>[[#References|1]]</sup>, but it was renamed to <i>K. oralis</i> in 1994<sup>[[#References|2]]</sup>. <i>K. oralis</i> is a gram-negative facultative (aerobic and anaerobic growth) organism thats grows in mesophillic environments. <i>K. oralis</i> is normally found in small numbers in the oral cavity in dental plaque, however periodontitis and gingivitis may correlate with increases in numbers of this species in the gingiva. They also produce corroding colonies. <i> K. oralis</i> is rather prevalent in dental plaque and has shown to constitute more than 5% of the total microbiota in peridontitis sites<sup>[[#References|3]]</sup>. | ||
==Genome structure== | ==Genome structure== | ||
<i>K. Oralis</i> strain ATCC 51147 does not encode for chromosomes and plasmids. It has a total length of about 2.41mb, encodes for 2,315 proteins and has a GC content of 54.3%. Sequencing results show that ATCC51147 consists of 5 scaffolds (Scfld 0-4) with no gaps between, an assembly gap length of 700, and 12 contigs. It has 3,165 coding genes and 52 non-coding genes. From a genome anotation, 6 rRNA (3 5S, 1 16S, 1 23S and 1 partial 23S), 48 tRNA and 2ncRNA sequences were identified | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
<i>K. oralis</i> are gram negative rods or cocobacilli that can form pairs or chains. It has a cell wall that consists of an outer membrane containing lipopolysaccharides, a periplasmic space with a peptidoglycan layer, and an inner, cytoplasmic membrane. Cells are nonmotile by flagella, but have monopolar fimbriae and form spreading colonies which suggests twitching motility<sup>1</sup>. <i>K. oralis</i> were not known to coaggregate, however recent studies have shown that they are able to coadhere with <i>Streptococcus oralis</i> during the formation of complex biofilms</i><sup>[[#References | 7]]</sup>. Cells can be aerobic or facultatively anaerobic. They are oxidase positive, catalase negative and are able to ferment glucose. | |||
==Ecology== | ==Ecology== | ||
<i>K. oralis</i> is an aerobic or facultatively anaerobic organism that is often cultured for on blood agar. The main habitat of <i>K. oralis</i> is in human dental plaque, however, it can also be found in smaller quantities in salival and mucosal sites. In periodontally healthy patients, <i>K. Oralis</i> is relatively evenly distributed in subgingival and supragingival plaque, but it has a higher distribution in the supragingival plaque of those with | <i>K. oralis</i> is an aerobic or facultatively anaerobic organism that is often cultured for on blood agar. The main habitat of <i>K. oralis</i> is in human dental plaque, however, it can also be found in smaller quantities in salival and mucosal sites. In periodontally healthy patients, <i>K. Oralis</i> is relatively evenly distributed in subgingival and supragingival plaque, but it has a higher distribution in the supragingival plaque of those with periodontitis <sup>[[#References|9]]</sup>. | ||
The Kingella species are known to exchange antibiotic resistance plasmids with <i>Neisseria gononhoeae</i> and <i>Neisseria | The Kingella species are known to exchange antibiotic resistance plasmids with <i>Neisseria gononhoeae</i> and <i>Neisseria meningitidis</i>, resulting in a possibility of <i>K. oralis</i> as a plasmid reservoir<sup>[[#References|10]]</sup>. | ||
meningitidis</i>, resulting in a possibility of <i>K. oralis</i> as a plasmid reservoir | |||
==Pathology== | ==Pathology== | ||
[[File:Gingivitis_and_periodontitis.jpeg|200px|thumb|left|]] | |||
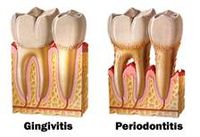
<i>K. oralis</i> is an organism that is capable of locally invading periodontal tissues and inducing host defense. This contributes to gum diseases such as gingivitis and periodontitis that affects both adults and juveniles<sup>[[#References|9]]</sup>. | |||
<i>K. oralis</i> is | Gingivitis is usually the precursor stage of gum disease and is caused by gum inflammation due to <i> K. oralis</i> and other bacteria accumulating as plaque or tartar on teeth. In gingivitis, the gums become red, swollen, and bleeds easily. However, with regular cleaning of teeth, gingivitis can usually be reversed. | ||
When gingivitis is left untreated, there is a possibility of periodontitis (inflammation around the tooth) occurring. This causes gums to pull away from the teeth to form pockets that become infected. Bacterial toxins produced combined with the body's immune response to the infection causes the breakdown of bone and connective tissue that hold the teeth in place. When left untreated, the teeth may eventually become loose and would have to be removed. | |||
However, by maintaining good oral hygiene and practices, gum diseases such as these can be controlled.<sup>[[#References|8]]</sup> | |||
==Application to biotechnology== | ==Application to biotechnology== | ||
<i>K. oralis</i> has been used in the validation of a new assay that can be used in the screening for coadhesion partners among the multitude of species present in oral biofilms<sup>7</sup>. | <i>K. oralis</i> has been used in the validation of a new assay that can be used in the screening for coadhesion partners among the multitude of species present in oral biofilms<sup>[[#References|7]]</sup>. | ||
There has been no other evidence that shows that this species has been used in biotechnology. | There has been no other evidence that shows that this species has been used in biotechnology. | ||
==Current research== | ==Current research== | ||
'''Probing of Microbial Biofilm Communities for Coadhesion Partners''' | |||
<i>K. oralis</i> was not known to coaggregate with other species during biofilm formation, however recent research investigating the interbacterial adhesion in dental plaque development has identified <i>K. oralis</i> to be able to coaggregate with <i>Streptococcus oralis</i> during the formation of complex biofilms<sup>[[#References|7]]</sup>. | |||
Another recent research on DNA Uptake Sequence (DUS) has discovered that <i>K. oralis</i> replication is DUS-dependent(kingDUS) and this affects transformation by limiting DNA uptake and recombination in favour of homologous DNA. Approximately 2.5% of the <i>K. oralis</i> genome is occupied by the kingDUS, and this is very high compared to the approximate 1% DUS occupancy in <i> | |||
'''Dialects of the DNA Uptake Sequence in <i>Neisseriaceae</i>''' | |||
Another recent research on DNA Uptake Sequence (DUS) has discovered that <i>K. oralis</i> replication is DUS-dependent(kingDUS) and this affects transformation by limiting DNA uptake and recombination in favour of homologous DNA. Approximately 2.5% of the <i>K. oralis</i> genome is occupied by the kingDUS, and this is very high compared to the approximate 1% DUS occupancy in <i>Neisseriaceae meningitidis</i> and <i>Neisseriaceae gonorrhoeae</i> genomes<sup>[[#References|6]]</sup>. | |||
==References== | ==References== | ||
1. [http://www.microbiologyresearch.org/docserver/fulltext/ijsem/43/3/ijs-43-3-490.pdf?expires=1474385696&id=id&accname=guest&checksum=2BD07E35E212EBADD9B3C356483EABE4 Dewhirst F, Chen C, Paster B, Zambon J. 1993. Phylogeny of Species in the Family Neisseriaceae Isolated from Human Dental Plaque and Description of Kingella orale sp. nov. International Journal of Systematic Bacteriology 43:490-499] | 1. [http://www.microbiologyresearch.org/docserver/fulltext/ijsem/43/3/ijs-43-3-490.pdf?expires=1474385696&id=id&accname=guest&checksum=2BD07E35E212EBADD9B3C356483EABE4 Dewhirst F, Chen C, Paster B, Zambon J. 1993. Phylogeny of Species in the Family Neisseriaceae Isolated from Human Dental Plaque and Description of Kingella orale sp. nov. International Journal of Systematic Bacteriology 43:490-499] | ||
| Line 60: | Line 66: | ||
3. [http://www.ncbi.nlm.nih.gov/pubmed/9467377 Chen C. 1996. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiology and Immunology 11:425-427] | 3. [http://www.ncbi.nlm.nih.gov/pubmed/9467377 Chen C. 1996. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiology and Immunology 11:425-427] | ||
4. [http://www.ncbi.nlm.nih.gov/genome/?term=Kingella%20oralis | 4. [http://www.ncbi.nlm.nih.gov/genome/?term=Kingella%20oralis NCBI-Genome] | ||
5. [http://www.antimicrobe.org/new/b66.asp Yagupsky, P. Kingella species. Antimicrobe] | 5. [http://www.antimicrobe.org/new/b66.asp Yagupsky, P. Kingella species. Antimicrobe] | ||
6 | 6. [http://journals.plos.org/plosgenetics/article?id=10.1371%2Fjournal.pgen.1003458 Frye S, Nilsen M, Tønjum T, Ambur O. 2013. Dialects of the DNA Uptake Sequence in Neisseriaceae. PLoS Genetics 9:e1003458] | ||
7. [http://aem.asm.org/content/80/21/6583.full Ruhl S, Eidt A, Melzl H, Reischl U, Cisar J. 2014. Probing of Microbial Biofilm Communities for Coadhesion Partners. Applied and Environmental Microbiology 80:6583-6590] | |||
8. [http://www.nidcr.nih.gov/oralhealth/Topics/GumDiseases/PeriodontalGumDisease.htm 2014. Periodontal (Gum) Disease: Causes, Symptoms, and Treatments. National Institute of Dental and Craniofacial Research] | |||
9. [http://onlinelibrary.wiley.com.ezproxy.library.uq.edu.au/doi/10.1111/j.1399-302X.1996.tb00206.x/epdf Chen C. 1996. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiology and Immunology 11:425-42] | |||
10. [http://aac.asm.org/content/32/5/765.short Knapp, J. S., S. R Johnson, J. M. Zenilman, M. C. Roberts, and S. A. Morse. 1988. High-level tetracycline resistance resulting from TetM in strains of Neisseria spp., Kingella denitrificans, and Eikenella corrodens. Antimicrob. Agents Chemother. 32:765-767] | |||
11. [http://www.beaversdentistry.com/cms/wp-content/uploads/2014/04/periodontistis.jpg Image-Gingivitis & Periodontitis] | |||
Latest revision as of 12:25, 23 September 2016
Esther Chua | Bench E | 31 Aug 2016 [1]
Classification
Higher order taxa
Bacteria - Proteobacteria - Betaproteobacteria - Neisseriales - Neisseriaceae - Kingella - Kingella oralis
Species
Kingella oralis
Type strain: strain UB-38 = ATCC 51147= CCUG 30450 = CIP 103803
Description and significance
Kingella oralis, K. oralis was first obtained from human periodontal isolates recovered from a purported Eikenlla corrodens-selective medium1. In 1990, it was originally described as Eikenella corrodens-like isolates2. Using 16sRNA sequencing, these isolates were identified as K. orale in 19931, but it was renamed to K. oralis in 19942. K. oralis is a gram-negative facultative (aerobic and anaerobic growth) organism thats grows in mesophillic environments. K. oralis is normally found in small numbers in the oral cavity in dental plaque, however periodontitis and gingivitis may correlate with increases in numbers of this species in the gingiva. They also produce corroding colonies. K. oralis is rather prevalent in dental plaque and has shown to constitute more than 5% of the total microbiota in peridontitis sites3.
Genome structure
K. Oralis strain ATCC 51147 does not encode for chromosomes and plasmids. It has a total length of about 2.41mb, encodes for 2,315 proteins and has a GC content of 54.3%. Sequencing results show that ATCC51147 consists of 5 scaffolds (Scfld 0-4) with no gaps between, an assembly gap length of 700, and 12 contigs. It has 3,165 coding genes and 52 non-coding genes. From a genome anotation, 6 rRNA (3 5S, 1 16S, 1 23S and 1 partial 23S), 48 tRNA and 2ncRNA sequences were identified
Cell structure and metabolism
K. oralis are gram negative rods or cocobacilli that can form pairs or chains. It has a cell wall that consists of an outer membrane containing lipopolysaccharides, a periplasmic space with a peptidoglycan layer, and an inner, cytoplasmic membrane. Cells are nonmotile by flagella, but have monopolar fimbriae and form spreading colonies which suggests twitching motility1. K. oralis were not known to coaggregate, however recent studies have shown that they are able to coadhere with Streptococcus oralis during the formation of complex biofilms 7. Cells can be aerobic or facultatively anaerobic. They are oxidase positive, catalase negative and are able to ferment glucose.
Ecology
K. oralis is an aerobic or facultatively anaerobic organism that is often cultured for on blood agar. The main habitat of K. oralis is in human dental plaque, however, it can also be found in smaller quantities in salival and mucosal sites. In periodontally healthy patients, K. Oralis is relatively evenly distributed in subgingival and supragingival plaque, but it has a higher distribution in the supragingival plaque of those with periodontitis 9.
The Kingella species are known to exchange antibiotic resistance plasmids with Neisseria gononhoeae and Neisseria meningitidis, resulting in a possibility of K. oralis as a plasmid reservoir10.
Pathology
K. oralis is an organism that is capable of locally invading periodontal tissues and inducing host defense. This contributes to gum diseases such as gingivitis and periodontitis that affects both adults and juveniles9.
Gingivitis is usually the precursor stage of gum disease and is caused by gum inflammation due to K. oralis and other bacteria accumulating as plaque or tartar on teeth. In gingivitis, the gums become red, swollen, and bleeds easily. However, with regular cleaning of teeth, gingivitis can usually be reversed.
When gingivitis is left untreated, there is a possibility of periodontitis (inflammation around the tooth) occurring. This causes gums to pull away from the teeth to form pockets that become infected. Bacterial toxins produced combined with the body's immune response to the infection causes the breakdown of bone and connective tissue that hold the teeth in place. When left untreated, the teeth may eventually become loose and would have to be removed. However, by maintaining good oral hygiene and practices, gum diseases such as these can be controlled.8
Application to biotechnology
K. oralis has been used in the validation of a new assay that can be used in the screening for coadhesion partners among the multitude of species present in oral biofilms7.
There has been no other evidence that shows that this species has been used in biotechnology.
Current research
Probing of Microbial Biofilm Communities for Coadhesion Partners
K. oralis was not known to coaggregate with other species during biofilm formation, however recent research investigating the interbacterial adhesion in dental plaque development has identified K. oralis to be able to coaggregate with Streptococcus oralis during the formation of complex biofilms7.
Dialects of the DNA Uptake Sequence in Neisseriaceae
Another recent research on DNA Uptake Sequence (DUS) has discovered that K. oralis replication is DUS-dependent(kingDUS) and this affects transformation by limiting DNA uptake and recombination in favour of homologous DNA. Approximately 2.5% of the K. oralis genome is occupied by the kingDUS, and this is very high compared to the approximate 1% DUS occupancy in Neisseriaceae meningitidis and Neisseriaceae gonorrhoeae genomes6.
References
4. NCBI-Genome
5. Yagupsky, P. Kingella species. Antimicrobe
11. Image-Gingivitis & Periodontitis
- ↑ MICR3004
This page is written by <Esther Chua> for the MICR3004 course, Semester 2, 2016