Vaginal Microbiota: Canal vs. C-Section
What is the Vaginal Mirobiota?

By Racine Ross
The vaginal microbiota consists of all the microorganisms that colonize the vagina but are also part of the general human microbiota. The vaginal microbiota is known to harbor over 250 species of bacteria such as Actinomyces, Bacteroides, Enterobacter, Mollicutes, Proteobacteria, Salmonella, and Streptococcus[1]. In 1892, Albert Döderlein became the first researcher to explain the significance of lactic acid-producing bacteria in the vaginal microbiome [2]. Since then, research has evolved to determine the most prominent species found in the vagina, the most common disturbance of the vaginal microbiome, the varying microbiota types, how the microbiota affects childbirth, and so much more. In normal function, the vaginal microbiota contains a mixture and balance of bacteria such as those previously mentioned. Although, comprehensive surveys of the vaginal microbiome have proved that Lactobacillus species are among the dominant vaginal bacterial species in a large proportion of women [3]. The action of ‘’Lactobacillus’’ in the vaginal microbiome is crucial to protecting women from genital infections as well as maintaining the natural balance of the vaginal microbiome. Not only does ‘’Lactobacillus’’ affect the vaginal microbiome of the mother, but it also impacts one of the first bacteria introduced to a fetus. More specifically, babies born vaginally were colonized by ‘’Lactobacillus’’, but those born by c-section were colonized by a mixture of pathogenic bacteria such as ‘’Staphylococcus’’ and ‘’Acinetobacter’’ [4]. There are microbes that “seed” the intestine during cesarean delivery and vaginal delivery which may lead to changes that affect the long-term colonization and alteration of immune development (10).
Lactobacillus
Lactobacilli have the following scientific classification:
Domain: Bacteria
Phylum: Bacillota
Class: Bacilli
Order: Lactobacillales
Family: Lactobacillaceae
Genus: Lactobacillus
Lactobacilli are gram-positive, rod-shaped, non-spore-forming bacteria that grow well in microaerophilic conditions [3]. As a gram-positive firmicute, Lactobacilli have a thick cell wall with a cell envelope consisting of glycosyl chains, an s-layer, peptidoglycan with teichoic acids, and a cell membrane containing membrane specific proteins. Additionally, Lactoabacilli are members of the lactic acid bacteria whose main end product of carbohydrate metabolism is the formation of lactic acid. Lactobacilli’s nutritional requirements are well represented by the habitat they reside in. These are rich in carbohydrate-containing-substrates that can be found on plants or material of plant origin, in fermented or spoiled food, or in association with the bodies of animals (7). Moreover, Lactoabacilli is a necessary factor in food production that requires lactic acid fermentation such as dairy products (yogurt and cheese), fermented vegetables (olives, pickles, and sauerkraut), fermented meats (salami), and sourdough bread. Lactobacilli is known to be anaerobic bacteria but can also be found in niches supplemented with oxygen (2). Another place Lactobacilli can be found are in the vaginal microbiome. In the vagina, Lactobacilli have the ability to produce hydrogen peroxide, but this presence has been found to be negatively associated with the formation of bacterial vaginosis which will be discussed later (2). Lastly, Lactobacilli alone contains over 170 different species, with its most common in the vaginal microbiota being Lactobacillus acidophilus which is the species I will be focusing on.
Genome of Lactobacillus acidophilus
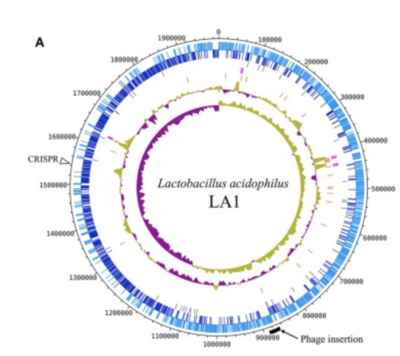
The complete genome is composed of a 1.99-Mbp circular chromosome with 34.7% G+C content, or the percentage of guanine-cytosine in the RNA (Figure 1). The genome of Lactobacillus acidophilus was sequenced and a total of 1,953 were found in the genome which included 1,844 protein-coding genes, 76 RNA genes, and 33 pseudogenes. There were 4 sets of ribosomal RNA genes which included 5S, 16S, and 23S genes. Additionally, 61 tRNA genes were found as well as three non-coding RNA [5].
A unique feature of the Lactobacillus acidophilus is a 26-kbp prophage insertion at the chromosomal position 863,940-890,001. This position and location are similar to other Lactobacillus acidophilus genes, with the only difference being the number of proteins encoded in that specific region [5]. LA1, a strain of Lactobacillus acidophilus, contains the smallest number of genes, and the genes required for insertion include attL, phage integrase, and attR [5].
Phylogenetic Comparisons
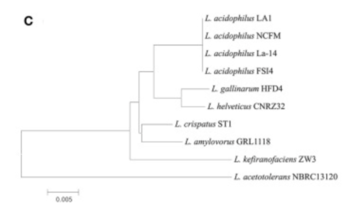

There appeared to be high conservation of the Lactobacillus acidophilus group. The similarity of the genome ranged from 75% to 99.9% for 10 complete genomes in this group. When analyzing similarities based on 16S rRNA, the range was from 94% to 100%. Despite the similarities present, researchers found differing phylogenetic relationships between the 16S rRNA sequence-based and average nucleotide identity value-based phylogenetic trees. Other Lactobacillus species such as Lactobacillus gallinarum and Lactobacillus helveticus were the closest to Lactobacillus acidophilus on the 16S rRNA phylogenetic tree (Figure 2)[5]. On the other hand, Lactobacillus amylovorus was the closest to Lactobacillus acidophilus on the average nucleotide identity phylogenetic tree (Figure 3)[5]. Studying phylogenetics using average nucleotide identity does a better job at reflecting the functional relationship between strains as opposed to studies that are based on 16S rRNA sequences. Lactobacillus gallinarum and Lactobacillus helveticus showed distinguishable profiles when compared to the Lactobacillus acidophilus genomes [5].
As compared to other species, Lactobacillus acidophilus had a lower G+C content. Logistically speaking, having a lower G+C content would mean that the bonds are less stable as opposed to a species that had a higher G+C content [5]. Gene clustering was also completed to understand the difference between 10 genomes that were based on the COG’s database functional annotation. From this database, the genes were categorized into 3,810 functional clusters. A total of 1,717 core gene clusters were found among the total 1,955 gene clusters. Despite the fact that there were high similarities among the Lactobacillus acidophilus genome, four of the gene clusters were found to be absent in the LA1 genome[5].
Energy Source
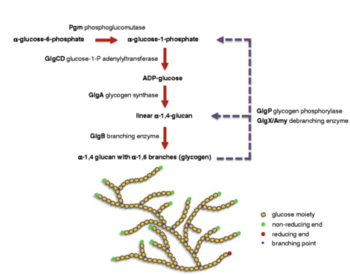
Lactobacillus acidophilus was the first probiotic microorganism demonstrated to possess a functional glycogen biosynthetic pathway [6]. It is known to be dependent on anaerobic respiration with byproducts including lactic acid from the catabolism of glucose and amino acids. Moreover, Lactobacilli use glucose as a carbon source and can either be homofermentative, meaning that they produce more than 85% of fermentative products as lactic acid, or they can be heterofermentative, meaning that they produce lactic acid, carbon dioxide, ethanol, and acetic acid in equimolar quantities [7].Interestingly enough, Lactobacillaceae are the only family of lactic acid bacteria that includes the two aforementioned organisms. In its metabolism process, Lactobacillus acidophilus uses the glycogen metabolic pathway. The pathway is encoded by an 11.7-kb chromosomal region that consists of 7 glycogen-specific Lactobacillus acidophilus genes. All seven of these genes are co-transcribed as a polycistronic mRNA and the gene cluster is designated as the glg operon [6]. Furthermore, this glg- encoding species primarily originated from mammalian hosts or natural environments.
Researchers have also found that glycogen metabolism is dependent not only on oxygen but also on the carbon source and growth phase. Due to the fact that there is a different expression of the glg operon and glycogen buildup in varying carbohydrate conditions, it shows that glycogen biosynthesis conducted by Lactobacillus acidophilus depends on the type of sugar substrates that are present [6]. A microarray study of carbohydrates used by Lactobacillus acidophilus was conducted which showed that the glg operon was upregulated when raffinose (trisaccharide) or trehalose (a sugar) was given as the sole carbon source as compared to other sugars. Additionally, a PCR experiment, and quantitative glycogen assay showed that raffinose was one of the sugar substrates examined which produces the highest level of the glg operon expression and intracellular glycogen accumulation. On the contrary, the researchers found that glucose appeared to repress the glg expression and biosynthesis. All of this is consistent with the identification of a catabolite response element located upstream of the glg operon. Furthermore, this shows that glycogen metabolism is affected by catabolite regulation [6].
Environmental Interactions
Issues with an imbalance of bacteria
Vaginal dysbiosis is defined as the disruption of the microbial community in the vagina and is typically associated with gynecological diseases such as bacterial vaginosis, sexually transmitted infections, and vulvovaginal candidiases [1][8]. Moreover, these disruptions can lead to pregnancy loss, preterm labor, and low conception rates [8]. It is essential for the vaginal microbiome to stay healthy an in balance to prevent any unwanted diseases. Thankfully with modern medicine, there are treatments and antibiotics that can be taken to provide a cure for these gynecological diseases.
Bacterial Vaginosis
Bacterial vaginosis can be described by a grey-white milky discharge, pH>4.5, a bad “fishy” smell, and at least 20% of “clue cells” which are defined as cells in the vagina that have a fuzzy appearance without sharp edges [1]. The presence of bacterial vaginosis has been associated with ‘’Gardnerella vaginalis’’ which is an anaerobic gram-variable non-spore-forming anaerobic bacteria. There are currently 4 different strains of ‘’Gardnerella vaginalis’’ with two of them known to produce bacterial vaginosis. Despite this, researchers have discovered that there is no single strain that produces bacterial vaginosis. Although, researchers have deduced that women who have a higher concentration of ‘’Gardnerella vaginalis’’ are more likely to develop bacterial vaginosis [1].
Lots of recent evidence suggests that bacterial vaginosis in pregnancy is associated with the risk of preterm birth as well as potential neonatal sequelae due to prematurity. Although there is no concrete evidence as to the mechanism by which bacterial vaginosis may lead to preterm birth, a possible explanation is that there are alterations present in the host defense mechanism, leading to intrauterine infection [9]. Moreover, researchers have proposed that women who are immunologically hyporesponsive or are less responsive to changes in the immune system, may not be able to control large bacterial attacks thus making them predisposed to increasing infection that will impact the fetus. On the contrary, women who have exaggerated cytokine production at the maternal-fetal interface may have an increased risk of preterm labor if the microorganisms are able to gain access to the choriodecidual space which is the layer surrounding the amniotic sac [9]. Unfortunately, it is not possible to understand what the causative agent is in bacterial vaginosis in women. This is due to the fact that Koch’s postulate has not been fulfilled because the etiologic agent is necessary and sufficient to cause the disease, but it should not be found in those without the disease (3). Much more extensive research is required to fully understand the mechanisms in which bacterial vaginosis interacts with the vaginal microbiome.
Vulvovaginal Candidiasis
Vulvovaginal candidiasis, more frequently referred to as a vaginal yeast infection, is caused by the fungus ‘’Candida albicans’’. ‘’Candida albicans’’ is a polymorphic yeast that can engage in yeast-to-yeast morphogenesis under favorable conditions (6). In the vaginal microbiome, ‘’Candida albicans’’ is known to co-colonize the vagina alongside ‘’Lactobacillus’’ and is commonly isolated from vulvovaginal candidiasis. Researchers have come up with some explanations that describe how ‘’Candida albicans’’ is capable of switching between colonizer and pathogen. Some of these are vaginal dysbiosis, expression of virulence factor, and production of proteolytic enzymes which resulted in vaginal immune toxicity. Additionally, it has been demonstrated that intraepithelial abrasions in patients experiencing vulvovaginal candidiasis had ‘’Candida albicans’’ hyphae (branches) along with the co-invasion of ‘’Gardnerella vaginalis’’ (6). This evidence portrays that morphological plasticity enables yeast-to-hyphae formation in ‘’Candida albicans’’ and the presence of bacterial vaginosis-associated bacteria could be the cause of vulvovaginal candidiasis. Moreover, the disruption of the vaginal microbiota may encourage the capabilities of the ‘’Candida’’ species to invade the epithelial cells. This invasion causes a myriad of cascade effects and essentially triggers severe vaginal inflammation (6). ‘’Candida albicans’’ is still considered the main causative agent for vulvovaginal candidiasis.
Differences in outcomes between Canal and C-section
Conclusion
The human vaginal microbiome is a complex collection of microorganisms. The relationship between the vaginal microbiome and the microbes is mutually beneficial for the host and has a great impact on their health and wellbeing. Being able to understand why a balance is needed or how slight changes in the concentration of certain microbes can contribute to unwanted gynecological diseases or sexually transmitted infections, is essential for maintaining a healthy bacterial community. Changes in the vaginal microbiota have been found to induce changes such as diabetes, pregnancy, immunodeficiency-allergic rhinitis, and continual vulvovaginal candidiasis [8]. Even though there are a large number of microorganisms in the vaginal microbiota, ‘’Lactobacillus’’, specifically ‘’Lactobacillus acidophilus’’ make up the largest proportion of microbes. Its role in the vaginal microbiome consists of protecting the woman from harboring diseases and ensuring that things remain in balance. Not only does “Lactobacillus” affect the host, but can also affect the immunity development of a fetus. More research is needed to fully understand all of the ins and outs of the vaginal microbiome and what makes it an especially unique and essential environment.
References
- ↑ 1.0 1.1 1.2 1.3 Mendling, W.: Vaginal Microbiota. Advances in experimental medicine and biology, 902, 83–93.
- ↑ Ma, B., Forney, L. J., & Ravel, J. : Vaginal microbiome: rethinking health and disease. Annual review of microbiology, 66, 371–389.
- ↑ 3.0 3.1 Goldstein, E. J., Tyrrell, K. L., & Citron, D. M. (2015). Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 60 Suppl 2, S98–S107.
- ↑ Neu, J., & Rushing, J.(2011). Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clinics in perinatology, 38(2), 321–331.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 Chung, W. H., Kang, J., Lim, M. Y., Lim, T. J., Lim, S., Roh, S. W., & Nam, Y. D. (2018). Complete Genome Sequence and Genomic Characterization of Lactobacillus acidophilus LA1 (11869BP). Frontiers in pharmacology, 9, 83.
- ↑ 6.0 6.1 6.2 6.3 Goh, Y. J., & Klaenhammer, T. R. (2014). Insights into glycogen metabolism in Lactobacillus acidophilus: impact on carbohydrate metabolism, stress tolerance and gut retention. Microbial cell factories, 13, 94.
- ↑ Tannock G. W. (2004). A special fondness for lactobacilli. Applied and environmental microbiology, 70(6), 3189–3194.
- ↑ 8.0 8.1 8.2 Chee, W., Chew, S. Y., & Than, L. (2020). Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microbial cell factories, 19(1), 203.
- ↑ 9.0 9.1 Donati, L., Di Vico, A., Nucci, M., Quagliozzi, L., Spagnuolo, T., Labianca, A., Bracaglia, M., Ianniello, F., Caruso, A., & Paradisi, G. (2010). Vaginal microbial flora and outcome of pregnancy. Archives of gynecology and obstetrics, 281(4), 589–600.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2022, Kenyon College