Venezuelan equine encephalitis virus: Difference between revisions
| Line 41: | Line 41: | ||
VEEV has a non-segmented 11.4 kb (+) sense ssRNA genome with a poly (A) tail. The genome contains two reading frames that encode for two different polyproteins. Two-thirds of the genome starting from the 5’ end encode for a polyprotein that contains the four nonstructural proteins nsP1, nsP2, nsP3, and nsP4 (viral RNA-dependent RNA polymerase) that are required for genome replication. The last 4 kb of the genome encodes for a polyprotein with the capsid and envelope proteins CP, pE2, 6K, and E1 [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TD4-4XG52JN-M&_user=7774802&_coverDate=11%2F05%2F2009&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_searchStrId=1456107458&_rerunOrigin=google&_acct=C000062877&_version=1&_urlVersion=0&_userid=7774802&md5=01974ce50749b0fe533002b9057c9825&searchtype=a]. | VEEV has a non-segmented 11.4 kb (+) sense ssRNA genome with a poly (A) tail. The genome contains two reading frames that encode for two different polyproteins. Two-thirds of the genome starting from the 5’ end encode for a polyprotein that contains the four nonstructural proteins nsP1, nsP2, nsP3, and nsP4 (viral RNA-dependent RNA polymerase) that are required for genome replication. The last 4 kb of the genome encodes for a polyprotein with the capsid and envelope proteins CP, pE2, 6K, and E1 [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TD4-4XG52JN-M&_user=7774802&_coverDate=11%2F05%2F2009&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_searchStrId=1456107458&_rerunOrigin=google&_acct=C000062877&_version=1&_urlVersion=0&_userid=7774802&md5=01974ce50749b0fe533002b9057c9825&searchtype=a]. | ||
<br><br> | <br><br> | ||
The viral RNA itself contains four conserved sequence elements (CSEs) which interact with various viral and host proteins that are involved with the replication, transcription, and packaging of viral RNA. A small non-translated region at the beginning of the 5’ end of the genome (CSE1) appears to fold up into a stem-loop structure that acts as a promoter for (+) RNA strand replication from a (-) RNA template. CSE2 is found near the 5’ end as well but it is within the coding region for nsP1 and may be a promoter for the synthesis of (-) strand template RNA from genomic (+) strand RNA. CSE4, located just upstream of the poly (A) tail on the 3’ end of the RNA appears to be involved in promoting the synthesis of template RNA as well. CSE3 is located between the ORFs and is required for transcription of subgenomic RNA (smaller sections of mRNA that encode for only part of the genome). Just upstream Near CSE4 there are also repeated sequence elements 40-60-nt in length that bind to host proteins involved in the translation of viral RNA [ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2762864/?tool=pubmed]. | The viral RNA itself contains four conserved sequence elements (CSEs) which interact with various viral and host proteins that are involved with the replication, transcription, and packaging of viral RNA. A small non-translated region at the beginning of the 5’ end of the genome (CSE1) appears to fold up into a stem-loop structure that acts as a promoter for (+) RNA strand replication from a (-) RNA template. CSE2 is found near the 5’ end as well but it is within the coding region for nsP1 and may be a promoter for the synthesis of (-) strand template RNA from genomic (+) strand RNA. CSE4, located just upstream of the poly (A) tail on the 3’ end of the RNA appears to be involved in promoting the synthesis of template RNA as well. CSE3 is located between the ORFs and is required for transcription of subgenomic RNA (smaller sections of mRNA that encode for only part of the genome). Just upstream Near CSE4 there are also repeated sequence elements 40-60-nt in length that bind to host proteins involved in the translation of viral RNA [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2762864/?tool=pubmed]. | ||
==Virion Structure of Venezuelan equine encephalitis virus== | ==Virion Structure of Venezuelan equine encephalitis virus== | ||
Revision as of 05:18, 10 September 2010
Venezuelan equine encephalitis virus (VEEV)
A Viral Biorealm page on the family Venezuelan equine encephalitis virus
Baltimore Classification
Group IV: (+) sense single-stranded RNA viruses
Higher Order Categories
Family: Togaviridae
Genus: Alphavirus
Complex: New World
Description and Significance
Venezuelan equine encephalitis virus (VEEV) is an important animal and human pathogen found only in the Americas (the New World). VEEV is actually a complex of 7 different species that also include multiple subtypes and varieties [4]. Depending on the subtype, VEEV maintains either an enzoonotic or epizootic life cycle. Enzoonotic species sustain a chain of transmission between a rodent reservoir and mosquito vector. Epizootic species cause epidemics in horses and other animals of agricultural importance. during epidemics VEEV is spread by different species of mosquito vector. Most human cases of VEEV result from spill-over into the human population from an epizoonotic outbreak. Human are only incidental and usually dead-end hosts.
In the 1960's VEEV was weaponized by the U.S. and the U.S.S.R.[5]. The virus is considered a category B select bioterrorism threat by the U.S. government because it causes only moderate morbidity and low mortality rates in humans.
The virus has a biosafety level rating of 3 (BSL 3) which means that it can only be worked with inside of specially equipped laboratories designed to deal with BSL 3 agents [6]. although VEEV is transmitted by mosquitoes in nature it is a very good aerosol. The virus has been implicated in more than 150 cases of accidental laboratory exposure that were not associated with cuts or needle pricks [7].
VEEV is an arbovirus (arthropod-virus), a general term used to describe viruses that are transmitted via hematophagous arthropod vectors such as mosquitoes, flies and ticks. To be considered a true arbovirus there must be replication of the virus within the vector and VEEV is no exception [8].
Within the classification of arboviruses there is the genus alphavirus that includes VEEV. Alphaviruses cause arthritic and encephalitic disease in mammals. This group includes the pathogens O'nyong'nyong virus, Chikungunya virus, Western equine encephalitis virus, and Eastern equine encephalitis virus. Alphaviruses are not just important human and animal pathogens but have also been used as a model system in the study of enveloped virus structure and as viral vectors in gene therapy research [9][10].
Genome Structure
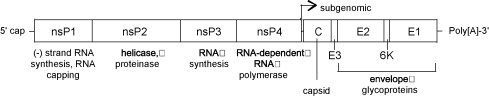
VEEV has a non-segmented 11.4 kb (+) sense ssRNA genome with a poly (A) tail. The genome contains two reading frames that encode for two different polyproteins. Two-thirds of the genome starting from the 5’ end encode for a polyprotein that contains the four nonstructural proteins nsP1, nsP2, nsP3, and nsP4 (viral RNA-dependent RNA polymerase) that are required for genome replication. The last 4 kb of the genome encodes for a polyprotein with the capsid and envelope proteins CP, pE2, 6K, and E1 [11].
The viral RNA itself contains four conserved sequence elements (CSEs) which interact with various viral and host proteins that are involved with the replication, transcription, and packaging of viral RNA. A small non-translated region at the beginning of the 5’ end of the genome (CSE1) appears to fold up into a stem-loop structure that acts as a promoter for (+) RNA strand replication from a (-) RNA template. CSE2 is found near the 5’ end as well but it is within the coding region for nsP1 and may be a promoter for the synthesis of (-) strand template RNA from genomic (+) strand RNA. CSE4, located just upstream of the poly (A) tail on the 3’ end of the RNA appears to be involved in promoting the synthesis of template RNA as well. CSE3 is located between the ORFs and is required for transcription of subgenomic RNA (smaller sections of mRNA that encode for only part of the genome). Just upstream Near CSE4 there are also repeated sequence elements 40-60-nt in length that bind to host proteins involved in the translation of viral RNA [12].
Virion Structure of Venezuelan equine encephalitis virus
Venezuelan equine encephalitis virus has an enveloped isohedral nucleocapsid. The capsid has a triangulation of t=4. Virions appear round and are 65-70 nm in diameter. The envelope surrounding the nucleocapsid (NC) is derived from host lipid bilayer embedded with approximately 240 copies of E1 and E2 glycoproteins. The glycoproteins form heterodimer spikes that link up in an isohedral lattice that also displays t=4 symmetry like the NC [ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2762864/?tool=pubmed].
Reproductive Cycle of Venezuelan equine encephalitis virus in a Host Cell
Little is specifically known about the KBV reproductive cycle. Some insight into the process can be gained by examining related viruses and examining the patterns of viral RNA and protein expression in infected bees.
The infective ability and life cycle of Rhopalosiphum padi virus (RhPV), also of the Cripavirus genus, has been studied to some extent. In cell culture, viral protein is localized to the cytoplasm, as visualized by immunogold labeling. This study also observed clusters of single membrane vesicles associated with virions in the early stages of infection. Crytalline arrays of virus particles and dense amorphous cytoplasmic structures were also observed.[3] Similar observations have been made for related viruses so it is reasonable to think that KBV infection may follow a similar scheme.
Another clue can be taken from the apparent ability of KBV to maintain a latent infection. Since KBV does not have a DNA intermediate, it cannot enter true lysogeny and splice into the host genome but it can remain in the cell without any observable pathology. Molecular evidence for latent infection was found when ELISA (immuno assay)results for viral-capsid proteins and RT-PCR amplification of viral RNA where compared as diagnostic techniques. In a majority of cases, viral RNA was present with little or no viral-capsid proteins.[9] This suggests that the viral RNA can persist in infected cells with limited replication and formation of new virus particles.
Viral Ecology & Pathology
animal hosts
skeeters are vectors-what species specifically
what it does-mostly flu-like illness, no biggie BUT can cause encephalitis and death
KBV is known to infect many species of bees across a large geographic distribution. Cases of A. melifera infection have been reported worldwide, although cases involved in CCD have only been reported in the US. KBV is also known to infect A. cerana (Asiatic honey bee) in parts of Southeast Asia and India, where the virus is believed to have originated. KBV has also been reported in populations of Bombus spp. (bumble bees) in New Zealand and Vespula germanica (European wasp) in Australia. [6] Some evidence shows that mites such as Varroa destructor not only act as vectors for viral sread, but also as hosts in which viral replication occurs [9].
KBV is spread between bees in a colony and between colonies through many methods of transmission. KBV can be introduced to hives by mites such as Varroa destructor [4] and can also be spread between worker bees and from the queen bee to her offspring. Viral RNA has been amplified from queens and their eggs suggesting that the virus can be transferred via a transovarial route. Viral RNA has also been amplified from food sources such as honey, brood food, and royal jelly. Since these food sources are partially composed of secretions from worker bees, this is a likely method of bee-to-bee transmission. [9]
As mentioned previously, KBV infection of shows no visually detectable signs of pathology [9]. KBV is commonly found in bees co-infected with other related viruses.
Vaccines and Treatments
Currently treatment of VEEV infection is mostly supportive because there are no specific drugs for alphaviruses but there is a vaccine for both humans and horse. The live attenuated vaccine known as TC-83 is a strain of VEEV that was passed 83 times in guinea pig heart cells (vaccine). There is also an inactivated form of the vaccine known as C-84 derived from the TC-83 strain. Currently only the C-84 vaccine is liscensed for us in horses in the U.S. although countries such as Mexico and Colombia still produce the live vaccine for horses. In the U.S. only at risk military and laboratory personal are vaccinated with the TC-83 strain and some recieve C-84 boosters if the intial vaccination did not produce sufficient immunity. The vaccine does have side affects that ranged from mild to moderate and did not provided full protection of nonhuman primates challenged by aersol exposure the route of transmission most likely if VEEV were to be used in a biological terrorist attack
References
[1] Jose, J., J.E. Snyder, R.J. Kuhn. “A structural and functional perspective of alphavirus replication and assembly.” Future Microbiology 4 (2009): 837-856
[2] Berenyi, O., T. Bakonyi, I. Derakhshifar, H. Koglberger, N. Nowotny. “Occurrence of Six Honeybee Viruses in Diseased Australian Apiaries.” Applied and Environmental Microbiology 72.4 (2006): 2412-2420.
[3] Boyapalle, S., N. Pal, W.A. Miller, B.C. Bonning. “A glassy-winged sharpshooter cell line supports replication of Rhopalosiphum padi virus (Dicistroviridae). Journal of Invertebrate Pathology 94 (2007): 130-139.
[4] Chen, Y.P., J.S. Pettis, A. Collins, M.F. Feldaufer. “Prevalence and Transmission of Honeybee Viruses.” Applied and Environmental Microbiology 72.1 (2006): 606-611.
[5] “Cripavirus.” ICTVdB Descriptions <www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.101.0.01.htm>
[6] de Miranda, J.R., M. Drebot, S. Tyler, M. Shen, C.E. Cameron, D.B. Stoltz, S.M. Camazine. “Complete nucleotide sequence of Kashmir bee virus and comparison with acute bee paralysis.” Journal of General Virology 85 (2004): 2263-2270.
[7] “Kashmir bee virus.” NCBI Taxonomy browser <www.ncbi.nlm.nih.gov/Taxonomy/Browser>
[8] Oldroyd, Benjamin P. “Unsolved Mystery: What’s Killing American Honey Bees?” PLoS Biology 5.6 (2007).
[9] Shen, M., L. Cui, N. Ostiguy, D. Cox-Foster. “Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite.” Journal of General Virology 86 (2005): 2281-2289.
[10] Slonczewski, J.L., J.W. Foster. “Chapter 6: Virus Structure and Function.” Microbiology: An Evolving Science (2009): 181-217.
Page authored for BIOL 375 Virology, September 2008
