A novel strain of avian origin influenza A (H7N9)
Introduction
By: Jack McDonald
Many of the infectious microbes that affect the world today have originated in animal hosts and made a host switch to infect humans. Although many of these diseases are able to affect humans, they are generally still able to infect their original host or a closely related microbe infects the original host. The majority of these infections are caused by viruses, bacteria or parasites. These types of infections include such illnesses as African trypanosomiasis (African sleeping sickness), malaria and many others. All of these diseases originated in an animal host and now infect both the animal of origin and humans alike. These diseases and infections are often transmitted through fluid exchange, contact with animal waste products or transmission through some other kind of vector (e.g. insects). One type of illness like this is caused by the influenza virus, in particular avian origin influenza. Influenza virus transmission is usually caused by direct contact with the disease vector or through contact with another infected individual. Avian origin influenza includes all H1 through H16 subtypes [3]. A novel strain of avian origin influenza A that has been recently discovered is the H7N9 strain. This particular strain first emerged in the spring of 2013 in northeastern China. It is of importance because it is the first documented occasion in which a human has been infected by an N9 type influenza virus [1]. The H7N9 strain has been classified by the CDC as a “low pathogenic avian influenza” (LPAI) because it is not seen to be exceedingly lethal to infected poultry [4]. However, this does not mean that its lethality is also diminished in humans. In fact, approximately one third of all cases of infection have ended in death [3]. As of now, the strain is unable to jump from human to human but development of this ability is still a possibility.
H7N9 Morphology
Influenza A viruses are a genus of the family orthomyxoviridae. The virus particle itself is spherical or oval in shape and is between 80 and 120 nanometers in diameter. The membrane of the virus particle is composed of a lipid bilayer, which is made from the cell membrane of the host cell from which it emerged. Embedded in the lipid membrane are glycoproteins called hemagglutinin and neuraminidase [6]. Hemagglutinin is a protein tetramer and is the part of the virus particle that attaches to the host cell to begin infection. Neuraminidase is believed to be the virus particle component that facilitates the emergence of new viral progeny from the host cell once infection is complete [6]. On the inside of the lipid membrane is the protein capsid, which houses the viral genome. The genome is composed of eight strands of single stranded RNA. The single strands of RNA are bound together in association with protein complexes to form helical ribonucleoproteins [5]. Each segment of the ribonucleoproteins has three different RNA polymerases associated with it (figure 1). Different strains of influenza virus are named according to the mutations they posses in relation to their hemagglutinin and neuraminidase in their membranes. For example, H7N9 has membrane glycoproteins of a specific structure and combination, which make it so that the human body is unable to effectively kill the virus particles using the normal immune response [6]. Also, mutations in the glycoproteins used for binding to potential host cells allow for virus particles to change which parts of a cell they can bind to. This leads to a kind of evolutionary arms race in which viruses and host organisms are constantly altering the structures of cell or particle components to ensure or prevent infection respectively.
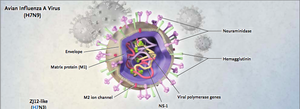
H7N9 Origins
As the name implies this particular strain of influenza virus is an avian origin virus. Most strains of avian flu are acquired through direct contact with poultry either in open markets or in agriculture. Poultry generally acquire the viral infection from other birds, usually wild, migratory, water dwelling birds. The genes responsible for the expression of the glycoprotein hemagglutinin (HA) have been linked to a strain of influenza isolated from domestic ducks in the Zhejiang province in China [2]. The gene responsible for the expression of the glycoprotein neuraminidase (NA) is similar to that of a strain of influenza isolated from wild, migratory ducks in eastern China. There were also some RNA polymerase genes that were similar to that of an avian influenza strain that has been isolated from bramblings, a migratory bird native to East Asia or possibly from other migratory birds from Europe [2]. These results indicate that this novel strain of avian origin influenza has been passed from migratory birds to species of birds that are commonly eaten and farmed by humans (figure 2). There are three different regional classifications of avian origin influenza and they are American, Eurasian and Oceanian. Complete genetic analysis indicates that the novel strain is of Eurasian origin [10]. Within these species the virus has mutated making it an effective pathogen to human beings. Evidence points to the fact that reassortment of the virus occurred in birds and not in humans [1]. The cases that have been confirmed have reported 75% of cases resulting from direct contact with poultry (mainly chickens), usually at open markets in cities. One theory states that there was an intermediate host in which the virus mutated from its original form in wild birds to a strain that was able to infect humans [2]. Although this particular strain is genetically unique to any existing strain of avian origin influenza found in China, it is genetically similar to other strains of avian flu found in other birds in eastern China.
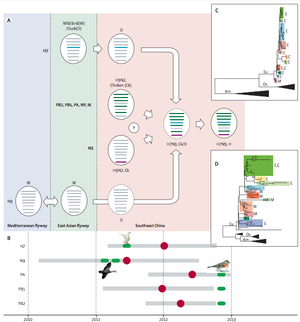
Pathology
The majority of influenza infections occur due to H1, H2 and H3 subtypes of the virus (common flu) [3]. Infections of the new strain of influenza (H7N9) have only been reported in eastern China. About 75% of those that have been infected have all had some kind of contact with poultry, either in a market setting or in the home. The mortality rate for this strain is approximately one third of total infections [3]. However, it is very likely that the mortality rate is actually much lower and that many of those that are infected are unaware that they are infected with the new strain and the infection goes unreported. Although this particular strain of influenza has caused death in humans, there has been no marked increase in poultry mortality as a result of the virus [3]. This is different than other strains of influenza, most notably the H5N1 strain, which caused increased mortality in both poultry and in humans. Because of the fact that H7N9 doesn’t cause increased mortality in poultry it could make the virus especially dangerous because detection and prevention in birds is much more difficult. Most strains of avian flu are very mild and result in pneumonia or conjunctivitis as symptoms of infection. This strain however has been shown to cause fever and lower respiratory tract infections of the lung epithelium, which causes acute respiratory distress syndrome and even death in extreme cases [3]. The average for onset of lung failure was between 3 and 14 days and those patients that did die died 1 to 3 weeks after the initiation of symptoms. Also, renal failure had been reported in at least one case of infection [1]. When this virus attacks cells it binds to sialic acid receptors to initiate the infection. In the epithelial tissue of the human respiratory tract there are α-2,6-linked sialic acid receptors [1]. The majority of infection from the H7N9 strain occurs in the lower respiratory tract, which causes acute respiratory distress syndrome [1]. Although the virus preferentially infects the lower respiratory tract, it does infect other areas such as the trachea, upper respiratory tract and even the brain [1]. To date there have been no confirmed instances of infection that have resulted from human-human contact. However, it has been demonstrated that the transmission of the H7N9 influenza strain can occur in ferrets (a model organism for human influenza infections) through direct contact and airborne transmission [7] (figure 3). In this particular study there were two groups of ferrets, a group inoculated with SH2 influenza (the novel H7N9 strain from Shanghai) and a group inoculated with CA07 (H1N1 strain from California). The two groups had three uninfected individuals placed in a cage with one infected individual (direct contact) and three individuals that had only airborne contact. In the SH2 group only one out of three ferrets were infected by airborne contact and all three were infected through direct contact. In the CA07 group all three ferrets in both the direct contact and airborne contact groups were infected with the virus [7]. This experiment is limited by the fact that ferrets and humans are not identical so findings associated with ferrets may not necessarily apply to humans, even though ferrets are the model organism for flu infections in humans. Also, another limitation of this experiment is its small sample size. To get a true indication of transmission it would be necessary to conduct a larger scale study. The findings of this experiment are still frightening because if the virus becomes able to be transmitted from human to human the virus could reach pandemic proportions and cause a large-scale outbreak. However, the fact that none of the reported infections have been confirmed to be derived from human-human contact is encouraging because it means that there will inherently be less infection than if it were possible to transmitted from human to human. So, perhaps spread of the virus due to direct contact or airborne transmission can only occur in other animals and not in humans. However, this leaves the possibility for the virus to further mutate allowing transmission from one human to the next. If this were to occur then the virus could reach pandemic proportions and kill millions of people worldwide.
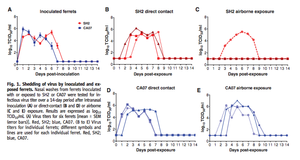
Avian flu comparison
Although the H7N9 strain is considered a novel strain of avian origin influenza virus, it is genetically and morphologically similar to a number of already existing strains. For example, the genes that code for polymerases inside the viral genome are very closely related to the genes that code for the same enzymes in an H9N2 strain [1]. This strain of influenza virus is mainly found in the area surrounding Beijing and in the city itself. The source of this strain is thought to be the aquatic bird called a brambling. However, it could also be possible that the similarity arose due to transport of poultry from Beijing to Shanghai [2]. This type of similarity between the strains likely does not make them similar in function. This is because the polymerase genes are involved in the replication of the genome and not in the pathogenic effects of the virus itself. The H7N9 strain also shares a similarity with another strain found in China, the H7N3 strain. These two both have an HA gene that is very similar to one another [3]. Because of this these two viral strains have very similar membrane glycoproteins that are associated with inducing binding to host cells. Because of this they likely initiate infection in a very similar way. This particular strain of avian influenza is not the first H7 subtype to infect humans and cause illness. There have been over one hundred different infections reported from across the world involving H7 subtype influenza viruses. One of these infections led to the death of a patient in the Netherlands [1]. One difference between other strains of avian flu, particularly H5N1 and other H7 strains, is that H7N9 has not shown to be readily transmitted from human to human [14]. This is good news for the time being because it means transmission of the virus is somewhat limited mostly to those that come into contact with poultry. Another difference between H7N9 and other influenza strains is that most avian flus are either asymptomatic or cause upper respiratory infection or conjunctivitis. H7N9 on the other hand causes lower respiratory tract infection that can lead to acute respiratory distress syndrome and renal and/or organ failure in some cases [7].
Outbreak history
The first confirmed infections of the new H7N9 strain of influenza occurred on March 30th, 2013 in Shanghai and the nearby province of Anhui [2]. The first infection was lab confirmed on March 30; however, the onset of symptoms for the first infected individual first occurred on February 18th [1]. Since the initial infections the occurrence of H7N9 was begun to spread throughout eastern and southern China and even into other southeastern Asian countries. For example, by the month of April, 2013 infection had spread to 6 other provinces and cities in China [2]. After one more month the total number of infections had risen to 125 confirmed human infections [7]. In addition to all of the confirmed infections in China there have been confirmed infections in other southeastern Asian countries including Malaysia and Taiwan. The spread of H7N9 has been slow to date and only progressed through a relatively small geographic area. This is likely due to the fact that the disease cannot be passed from one human to another or at least is not readily transmitted through human contact. However, if the virus does develop the ability to move from human to human the virus would most likely spread faster and further.
Potential risk factors
A common theme of infection with the H7N9 strain is that those who were infected tended to be older in age and many had other preexisting medical conditions. For example, in one study the mean age of the patients was 50 years old and all of the patients had some type of chronic ailment before they were infected with influenza virus [1]. Their conditions ranged from as mild as depression to as serious as hepatitis B infection [1]. This suggests that those that have their health compromised due to some preexisting condition are more likely to be infected by H7N9. Also, the fact that the mean age of infection is so old points to the virus infecting older individuals in the population. Establishing some kind of trend like this could aid health organizations in containing and helping to prevent or treat infections in the human population
Detection and treatment
Detection of a virus can be difficult in some cases because they can be hard to culture. This can make it difficult to get a big enough sample to obtain the necessary amount of material to determine if infection with a particular virus has occurred. The structure of the influenza virus makes it relatively easy to detect however. There are a number of different assays that can be done to determine the strain of influenza in question. For example, influenza virus has proteins associated with its genome called nucleoproteins (NP) and nucleoproteins are unique between strains allowing differentiation between the many strains [5]. Also, the structures of the glycoproteins in the lipid membrane of the virus particle are unique between strains, which also allow differentiation between strains [5]. The last common way that strains of influenza virus are detected and differentiated is through real time polymerase chain reaction (RT-PCR) in which the RNA genome of the virus is amplified and then can be sequenced [7]. If there is too small an amount of the virus then the virus particles can be cultured inside the yolks of chicken eggs to obtain the necessary amount of virus for analysis and classification. All of these techniques allow researchers and health care providers with viable and diverse ways to not only detect the presence of influenza virus, but also determine which strain of the virus is causing infection. Individuals that are infected by H7N9 influenza had a number of treatments that were performed to aid in recovery [1] (figure 4). Because of there decreased lung capacity and the onset of acute respiratory distress syndrome all patients were placed in some type of oxygen delivery or some other type of breathing support. In addition to this patients are administered some type of antiviral medication, usually oseltamivir [3]. In addition to antiviral medications some patients required intravenous immunoglobulin [3]. Immunoglobulin is an antibody, which is used to increase the body’s immune response and increase the body’s ability to effectively kill virus particles. Some studies have also found that the use of intravenous corticosteroids helped to fight the viral infection and promote recovery in infected patients [3]. There are many treatment options available for this novel strain of influenza that are effective and increase the chances of recovery. However, these treatment options are, for the most part, only available in modernized hospitals, which could be a problem if this virus became a pandemic. A vaccine has also been developed by the Chinese, which could help to quell any kind of large outbreak that may arise and maybe even prevent any such outbreak from occurring [8].
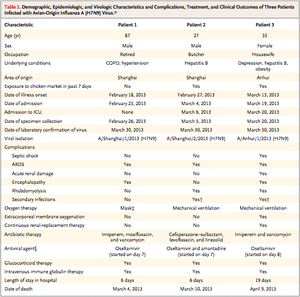
Conclusions and future impacts
Pathogens are constantly evolving and mutating so that they can infect new organisms or more effectively infect the organisms they already use as hosts. This is what has happened with the H7N9 strain of avian origin influenza A. The virus mutated and reassorted itself to host switch from its normal avian hosts to humans. This has rather serious implications from a global perspective because of the fact that a serious and deadly disease like this strain of influenza could potentially reach pandemic proportions with the right mutation. Most likely the only reason the virus hasn’t reached this level at this point is because human-human transmission does not occur or is very limited. If it does occur that H7N9 develops the ability to move from human to human the world could have another pandemic as serious as the 1918 Spanish flu outbreak. In today’s day and age it is likely that the death toll would be even higher simply due to the fact that the world population is so much higher now than it was in 1918. However, since there is a vaccine that has been developed for the H7N9 influenza strain, the spread of the virus may be stopped. A vaccine would allow China and other southeast Asian countries to offer the vaccine in their hospitals. This would allow the human population to stop the virus at its source and stop the global spread of the virus. Another positive aspect of the development of a vaccine for H7N9 is that even if there were to be a large outbreak due to human-human transmission the vaccine would be able to immunize those that are not yet infected and possibly help those that have been infected. The H7N9 strain has the potential to become a hugely devastating virus throughout the world. However, if steps are taken to fight and study the virus it is possible and even very likely that H7N9 can become a non-issue for Asian countries and the rest of the world.
References
[1] Gao, Rongbao, Bin Cao, Yunwen Hu, Zijian Feng, Dayan Wang, Wanfu Hu, Jian Chen, Zhijun Jie, Haibo Qiu, Ke Xu, Xuewei Xu, Hongzhou Lu, Wenfei Zhu, Zhancheng Gao, Nijuan Xiang, Yinzhong Shen, Zebao He, Yong Gu, Zhiyong Zhang, Yi Yang, Xiang Zhao, Lei Zhou, Xiaodan Li, Shumei Zou, Ye Zhang, Xiyan Li, Lei Yang, Junfeng Guo, Jie Dong, Qun Li, Libo Dong, Yun Zhu, Tian Bai, Shiwen Wang, Pei Hao, Weizhong Yang, Yanping Zhang, Jun Han, Hongjie Yu, Dexin Li, George F. Gao, Guizhen Wu, Yu Wang, Zhenghong Yuan, and Yuelong Shu. "Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus." New England Journal of Medicine 368.20 (2013): 1888-897. The New England Journal of Medicine. The New England Journal of Medicine. Web. 28 Mar. 2014. <http://www.nejm.org/doi/full/10.1056/NEJMoa1304459>.
[2] Liu, Di, Weifeng Shi, Yi Shi, Dayan Wang, Haixia Xiao, Wei Li, Yuhai Bi, Ying Wu, Xianbin Li, Jinghua Yan, Wenjun Liu, Guoping Zhao, Weizhong Yang, Yu Wang, Juncai Ma, Yuelong Shu, Fumin Lei, and George F. Gao. "Origin and Diversity of Novel Avian Influenza A H7N9 Viruses Causing Human Infection: Phylogenetic, Structural, and Coalescent Analyses." The Lancet 381.9881 (2013): 1926-932. Science Direct. The Lancet. Web. 28 Mar. 2014. <http://www.sciencedirect.com/science/article/pii/S0140673613609381>.
[3] Chen, Yu, Weifeng Liang, Shigui Yang, Nanping Wu, Hainv Gao, Jifang Sheng, Hangping Yao, Jianer Wo, Qiang Fang, Dawei Cui, Yongcheng Li, Xing Yao, Yuntao Zhang, Haibo Wu, Shufa Zheng, Hongyan Diao, Shichang Xia, Yanjun Zhang, Kwok-Hung Chan, Hoi-Wah Tsoi, Jade Lee-Lee Teng, Wenjun Song, Pui Wang, Siu-Ying Lau, Min Zheng, Jasper Fuk-Woo Chan, Kelvin Kai-Wang To, Honglin Chen, Lanjuan Li, and Kwok-Yung Yuen. "Human Infections with the Emerging Avian Influenza A H7N9 Virus from Wet Market Poultry: Clinical Analysis and Characterisation of Viral Genome." The Lancet 381.9881 (2013): 1916-925. Science Direct. The Lancet. Web. 28 Mar. 2014. <http://www.sciencedirect.com/science/article/pii/S0140673613609034>.
[4] "Avian Influenza A (H7N9) Virus." Centers for Disease Control and Prevention. Centers for Disease Control and Prevention, 12 Feb. 2014. Web. 4 Apr. 2014. <http://www.cdc.gov/flu/avianflu/h7n9-virus.htm>.
[5] Noda, Takeshi, Hiroshi Sagara, Albert Yen, Ayato Takada, Hiroshi Kida, R. Holland Cheng, and Yoshihiro Kawaoka. "Architecture of Ribonucleoprotein Complexes in Influenza A Virus Particles." Nature439.7075 (2006): 490-92. Nature. Nature. Web. 4 Apr. 2014. <http://www.nature.com/nature/journal/v439/n7075/abs/nature04378.html>.
[6] "Molecular Expressions Cell Biology: The Influenza (Flu) Virus." Molecular Expressions Cell Biology: The Influenza (Flu) Virus. Florida State University, n.d. Web. 4 Apr. 2014. <http://micro.magnet.fsu.edu/cells/viruses/influenzavirus.html>.
[7] Zhu, H., D. Wang, D. J. Kelvin, L. Li, Z. Zheng, S.- W. Yoon, S.- S. Wong, A. Farooqui, J. Wang, D. Banner, R. Chen, R. Zheng, J. Zhou, Y. Zhang, W. Hong, W. Dong, Q. Cai, M. H. A. Roehrl, S. S. H. Huang, A. A. Kelvin, T. Yao, B. Zhou, X. Chen, G. M. Leung, L. L. M. Poon, R. G. Webster, R. J. Webby, J. S. M. Peiris, Y. Guan, and Y. Shu. "Infectivity, Transmission, and Pathology of Human-Isolated H7N9 Influenza Virus in Ferrets and Pigs." Science 341.6142 (2013): 183-86.Science. Science. Web. 6 Apr. 2014. <http://www.sciencemag.org/content/341/6142/183.full.pdf>.
[8] "Influenza A Virus Subtype H7N9." Wikipedia. Wikimedia Foundation, 16 Apr. 2014. Web. 9 Apr. 2014. <http://en.wikipedia.org/wiki/Influenza_A_virus_subtype_H7N9>.
[9] "Avian Influenza A (H7N9) Virus." | Flu.gov. Flu.gov, n.d. Web. 9 Apr. 2014. <http://www.flu.gov/about_the_flu/h7n9/>.
[10] Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18(15):pii=20453. Available online: http://www.eurosurveillance.org/ViewArticle. aspx?ArticleId=20453
[11] Stevens, J., O. Blixt, J. C. Paulson, and I. A. Wilson. "Structure and Receptor Specificity of an Avian Flu Antigen." Structure and Receptor Specificity of an Avian Flu Antigen. National Institutes of Health and Skaggs Institute for Chemical Biology, n.d. Web. 03 May 2014. <http://www-als.lbl.gov/index.php/holding/210-structure-and-receptor-specificity-of-an-avian-flu-antigen.html>.
[12] "1918 Flu Pandemic." Wikipedia. Wikimedia Foundation, n.d. Web. 01 May 2014. <http://en.wikipedia.org/wiki/1918_flu_pandemic>.
[13] "H5N1 Avian Flu (H5N1 Bird Flu)." H5N1 (Avian Flu). N.p., n.d. Web. 03 May 2014. <http://www.flu.gov/about_the_flu/h5n1/>.
[14] Korteweg, Christine. "Pathology, Molecular Biology, and Pathogenesis of Avian Influenza A (H5N1) Infection in Humans." National Center for Biotechnology Information. U.S. National Library of Medicine, May 2008. Web. 03 May 2014. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2329826/>.
