Acinetobacter baumannii
A Microbial Biorealm page on the genus Acinetobacter baumannii
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter
Species
|
NCBI: Taxonomy |
Acinetobacter baumannii
Description and significance
Acinetobacter baumannii, previously named Acinetobacter calcoaceticus, is an opportunistic pathogen found in soil and water. One of the first Acientobacter found in soil was discovered in 1911 by M.W. Beijerinck. In the 1970s A. baumannii was susceptible to common antibiotics, but has now developed into a multidrug-resistant bacteria, capable of acquiring resistance genes. One of the first antibiotic resistant strains of A. baumannii called carbapenem-resistant A. baumanni (CRAB) was isolated in 1991. [1] A. baumannii generally affects patients with low immune systems, which has caused nosocomial infections and major concerns in hospitals given the ability of A. baumannii to live on a variety of hospital surfaces such as surgical drains and catheters. [2] Recently, there has been a growing number of blood stream infections caused by multidrug resistant A. baumanni among service members of the Iraq and Afghanistan military operations: Operation Iraqi Freedom and Operation Enduring Freedom, respectively. [3] Bloodstream infections caused by A. baumanni include pneumonia, urinary tract infections, and septicaemia. [4] Since A. baumannii has become more difficult to treat due to its increasing resitance to antibiotics such as carbapenem, beta-lactam, and tetracycline understanding its genomic sequence is essential to effectively treat infections and preventing A. baumannii transfer and growth on hospital surfaces.
Genome structure
A. baumannii is characterized by a single circular chromosome that contains 3,976,747 base pairs in which 3,454 are used for protein coding. [5] One strain of A. baumannii called AYE contains an 86kb resistance island, called AbaR1, that is made up of 45 resistance genes and is currently the largest island known to date. [6] A resistance island is a section on a chromosome that contains genes necessary to code for antibiotic resistance. Of those 45 resistance genes, 25 genes code for resistance against many antibiotics such as: tetracycline, aminoglycosides, cotrimoxazole, and chloramphenicol. [6] Not only does the resistance island code against antibiotics, but also for operons for arsenic and mercury resistance. [6] There are 14 resistance genes that code for class 1 integrons, which are sections of the chromosome capable of recombination, expression, and integration. [6] Mobility elements, such as transposase were found on 22 ORFs (open reading frames). [6] The A. baumanni AYE has three plasmids, but none contain resistance markers. [6] Not only does the strain AYE contain resistance genes, but also a common amino acid sequence with other organisms, which demonstrates genetic exchange, where “39 genes (44%) are likely to have originated from Pseudomonas spp., 30 (34%) from Salmonella spp., 15 (17%) from Escherichia spp., and four (4%) from other microorganisms”[6]
Cell structure and metabolism
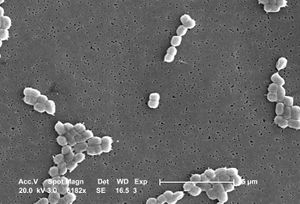
A. baumannii is an encapsulated Gram-negative coccobacilli bacteria that is generally non-motile. [4] Features on the outer cell membrane include porins and efflux channels which contribute to antibiotic resistance. In general, porins are protein channels that allow the transport of molecules across the membrane and are also sites of attachment for antibiotics. However, A. baumannii has fewer and smaller porins than other Gram-negative bacteria, thereby decreasing cell permeability and increasing antibiotic resistance. [7] Interestingly, less than 5% of molecules are permeable to the cell membrane, which is less than that found in Escherichia coli. [7] In some studies it was found that resistance to the antibiotic, carbapenem, was the result of the lack of a 29kDa outer membrane protein gene called carO. [8] It is believed that the protein CarO is involved with the import of carbapenem into the cell. [8]
Efflux pumps located in the cell membrane are used to pump chemicals and antibiotics out of the cell. Efflux pumps in A. baumannii include resistance to tetracycline called Tet (A) and Tet (B), part of the major facilitator superfamily (MFS) and functions in the exchange of protons and tetracycline. [7] There is also resistance-nodulation-cell division (RND) efflux pumps found in the strain A. baumannii BM4454 that is encoded by the gene adeB which provides aminoglycoside resistance. [9]
The cell wall on A. baumannii is not static, but rather changes in response to environmental conditions. In one study it was discovered that when placed in dry conditions there was an increase in the thickness of the cell wall, caused by a change in distance of the outer membrane and plasma membrane. [10] Also, there was a change in the shape of A. baumannii from rod shaped to cocci through a decrease in cell division. [10]
A. baumannii is a glucose non-fermentative, oxidase negative, aerobic bacteria. [4] Antibiotic resistant strains of A. baumannii produce beta-lactamases which can prevent antibiotic function by hydrolyzing penicillins, cephalosporins, and carbapenems. One of the first beta-lactamases, called ARI-1, but later changed to OXA-23 was collected in 1985 in Scotland, which hydrolyzed carbapenem. [4] Other OXA-type beta-lacamases include OXA-24 and OXA-58. [4]
Ecology
A. baumannii is able to survive on various surfaces in the hospital that are abiotic, wet, or dry. A. baumannii has the ability to avoid desiccation for more than 30 days on dry surfaces, although survival rate depends on where the specific strain originally came from (e.g. wet or dry conditions). [10]
One strain of A. baumannii, 19606 is capable of forming biofilm on glass and plastic surfaces via pili formation. [12] The production of biofilm may explain how A. baumanni can survive in different types of conditions in the hospitals, including static conditions such as bed sheets and furniture, while also capable of living in harsh conditions such as catheters and respiratory tubes. [12] A. baumannii also produces exopolysaccharides which strengthens the biofilm. [12]
Pathology
Acinetobacter baumannii causes 2-10% of all Gram-negative infections in the U.S. and Europe, poses little risk to healthy individuals, but generally causes infections to those with weakened immune systems. [6] Specifically, the intensive care unit (ICU) in hospitals houses patients with susceptible immune systems and is normally equipped with ventilators and invasive equipment such as catheters, that are factors that can contribute toA. baumannii infections such as pneumonia, meningitis, septicemia, and urinary and respiratory tract infections. In one study, it was shown that A. baumannii could cause apoptosis or cell death to human laryngeal epithelial cells via an outer membrane protein (OMP 38). [11] OMP38 releases cytochrome c and an apoptosis-inducing factor which enters the epithelial cell nucleus and causes the degradation of DNA (180bp). [11]
The ability of A. baumannii to cause untreatable infections is due to a variety of antibiotic resistance genes and cell surface structures that prevent the influx of antibiotics (see cell surface structure and metabolism). Since A. buamannii is commonly found in hospitals which is an environment where “antibiotics are frequently used, possessing integrons with multiple resistance determinants confers a strong selective advantage.” [3]
Application to Biotechnology
A. baumannii does not produce any enzymes for biotechnology.
Current Research
1. A. baumannii is easily transferred from not only patient to patient contact but also patient and environment contact. The study entitled “Importation of Acinetobacter baumannii Into a Burn Unit: A Recurrent Outbreak of Infection Associated With Widespread Environmental Contamination,” documents a patient infected with A. baumannii while being transported to the hospital and later infecting two other patients. What is mysterious is that the same strain that infected the first patients returned six months later in other wounded patients, despite the fact that tests were done after the first outbreak that showed that there were no other A. baumannii present. It is not clear how the A. baumannii strain reemerged after six months. [13]
2. In the study entitled, “Resistance to disinfectants in epidemiologically defined clinical isolates of Acinetobacter baumannii ,” researchers examined whether there was a correlation between the spread of A. baumannii and its reception to disinfectants. Researchers tested both sporadic and nosocomial strains and found that disinfectants worked well only if used properly with correct exposure time and not diluted. Since strict adherence is needed for disinfectants to be used well against A. baumannii, it may explain how the bacteria is capable of spreading and infecting individuals despite the use of disinfectants. [14]
3. Due to the growing number of A. baumannii antibiotic resistant bacteria and the lack of new forms of antibiotics to treat infections, the study entitled "In vitro activities of carbapenem/sulbactam combination, colistin, colistin/rifampicin combination and tigecycline against carbapenem-resistant Acinetobacter baumannii" examines the affects of combining different types of currently used antibiotics. Overall, the study showed that when culturing A. baumannii in the lab, combinations of antibiotics such as imipenem/sulbactam and colistin/tigecycline, proved effective against treating carbapenem resistant A. baumannii.[15]
References
[5] A. baumannii genome sequence: http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=search&db=genome
Edited by student of Rachel Larsen and Kit Pogliano