Alcaligenes ammonioxydans
Classification
Higher order taxa
- Domain: Bacteria
- Phylum: Proteobacteria
- Class: BetaProteobacteria
- Order: Burkholderiales
- Family: Alcaligenaceae
- Order: Burkholderiales
- Class: BetaProteobacteria
- Phylum: Proteobacteria
Species
Species
Alcaligenes ammonioxydans sp. nov. strain HO-1
Description and significance
The genus Alcaligenes is composed of Gram negative, typically aerobic species that have a wide range of metabolic capabilities, which included heterotrophic nitrification-aerobic denitrification (Hendrie et al. 1974; Wu et al. 2021). Alcaligenes ammonioxydans sp. nov. HO-1 was first isolated from the activated sludge of a single reactor system for high activity ammonium removal over nitrite (SHARON) bioreactor of a combined system which was used to treat wastewater from pig husbandry (Wu et al. 2021) This species is capable of high amounts of ammonia removal and HO-1 was capable of removing 195.59 mg/L NH4 per day on media supplemented with 28mM NH4. A. ammonioxydans grew aerobically and achieved ammonia removal on a range of carbon substrates which included acetate, ethanol, propionate, malate, succinate, and citrate. C/N ratios ranged from 2-20. Optimal pH is 7 but growth was observed at range of 6-10. Optimal temperature is 30°C with growth seen at 23-37°C but growth halts at 45°C (Wu et al. 2021).
16S Ribosomal RNA Gene Information
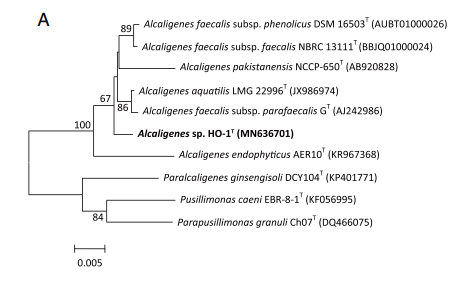
Alcaligenes ammonioxydans forms a cluster with other Alcaligenes isolates that is clearly separate from Alcaligenes faecalis but is still closely related (Wu et al. 2021)
Genome Structure
The complete genome of strain HO-1 is fully sequenced and consists of one chromosome and is composed of 3.77 Mbp with a DNA G+C content of 57.17%. The genome contained 3,320 predicted proteins with 57 tRNA sequences and 9 rRNA operons. Some genes were acquired horizontally as evident by 6 prophage sequences and 6 genomic islands. 80 tandem repeats and 63 interspersed repeats are found. Average nucleotide identity (ANI) analysis of related Alcaligenes genomes indicate that strain HO-1 belongs to a new, undescribed species (Wu et al. 2021).
Cell structure and metabolism
Cells are Gram-negative and are short rods with an average size of 0.8 µm in width and 1.0-1.5 µm in length. Strain HO-1 was isolated for its heterotrophic ammonia oxidation capabilities, and a novel metabolic pathway that involves the direct oxidation of ammonia to dinitrogen gas through a hydroxylamine intermediate was found. This novel pathway was called direct ammonia oxidation (Dirammox) and is translated from the novel gene cluster dnf. Genes dnfA, dnfB, and dnfC are all upregulated when ammonia is added to the culture and is hypothesized to play an important role in this pathway. Alcaligenes have a wide range of nitrogen metabolism capabilities. In addition to the dnf cluser, strain HO-1 contains genes that encode a complete denitrification pathway from nitrite (nirK, norBC, nosZ) as nitrite, but not nitrate, was shown to be reduced in anaerobic conditions. In aerobic conditions, neither nitrite nor nitrate was utilized as a sole nitrogen source. Thus, ammonia appears to be its main nitrogen source as the genome contains many ammonia transportation and assimilation genes. However, no genes encoding protein homologs of ammonia monooxygenase and hydroxylamine oxidoreductase were found (Wu et al. 2021).
Ecology
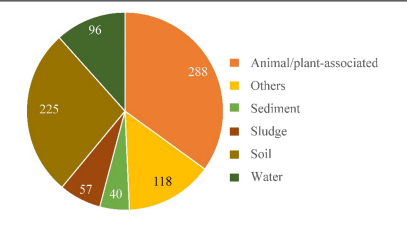
Strain HO-1 was isolated from a SHARON bioreactor that treats pig husbandry wastewater. Due to its high ammonia tolerance (up to 3,400 mg/L NH4) it is hypothesized to be a potential wastewater treatment strain (Hou et al. 2022). As strain HO-1 is a novel species, its distribution in the environment is not reported, but closely related Alcaligenes species were investigated based on 16S rRNA gene sequences in DNA databases. This analysis showed that Alcaligenes sequences were predominantly recovered in animal/plant associated samples (35% of samples) and in soil samples (27.3%). Sequences were also recovered in water (11.7%), sludge (6.9%) and sediment (4.85%). Since the genus Alcaligenes are widely distributed in environments that supports high levels of biodiversity it is hypothesized that species of Alcaligenes plays an important role in biogeochemical cycling, specifically through the nitrogen cycle with its Dirammox pathway. This is further supported by the fact that 31% of Alcaligenes strains were derived from wastewater treatment plants, agricultural wastewater, and sites impacted by various industrial effluents that contain high levels of nutrients (Hou et al. 2022).
Direct Ammonia Oxidation and Other Current Research
The novel direct ammonia oxidation (Dirammox) pathway was first discovered in Alcaligenes ammonioxydans HO-1 and it directly converts ammonia to dinitrogen gas via hydroxylamine under aerobic conditions. The dinitrogen gas produced was shown to be from this pathway and not that of nitrite denitrification (Wu et al. 2021. Through analysis of the 49 Alcaligenes genomes available in GenBank (as of May 2015), the dnf gene cluster was found in 36 genomes in the same orientation as found in strain HO-1. Of these genomes, A. faecalis was the most representative with 29 genomes, followed by unnamed strains with 4 genomes, and lastly A. aquatilis with 3 genomes. The dnf cluster of Alcaligenes spp. had a similar G+C content (56.3%) as that of whole Alcaligenes genomes (~57.2%) which indicates that the evolution of the dnf cluster was associated with members of the genus Alcaligenes. Thus, the dnf cluster was obtained from hereditary and not from horizontal gene transfer (Hou et al. 2022).
Dirammox is performed by enzymes encoded in the gene cluster dnfT1RT2ABCD, but just dnf ABC could enable a recombinant Escherichia coli strain to produce hydroxylamine and dinitrogen gas from ammonia (Wu et al. 2021). The seven genes of this cluster have been annotated to be 3-phosphoserine/phosphohydroxythreonine transaminase (T1) , PLP-dependent aminotransferase family protein (R), serine hydroxymethyltransferase (T2), diiron N-oxygenase (A), iron–sulfur cluster binding domain-containing protein (B), glutamine amidotransferase (C), and pyridoxine/pyridoxal/pyridoxamine kinase (D) (Wu et al. 2021). dnf R was shown to play a regulatory role and dnf A is crucial to the reaction as its deletion resulted in total loss of Dirammox activity in A. faecalis JQ135. As a result, dnfA was selected as a marker gene of the Dirammox pathway. Due to the general dinitrogen gas production of Alcaligenes and their ubiquity in nature, it is speculated that Dirammox is an important process in the nitrogen cycle, and its contribution must be assessed in situ (Hou et al. 2022).
Strain HO-1 was shown to have an antagonistic interaction with Bacillus velezensis and inhibited its growth through the accumulation of extracellular hydroxylamine. In B. velezensis, autolysis associated genes were upregulated in presence of hydroxylamine and genes involved in motility, chemotaxis, sporulation, polypeptide synthesis, and non-ribosomal peptide synthesis were all significantly downregulated. This indicates that HO-1 produces hydroxylamine as a byproduct of its metabolism which antagonizes surrounding bacteria while potentially acting as an interspecies signaling molecule (Gao et al. 2022). A new application of microbial biotechnology is using a metal based electrode as a final electron acceptor to support respiration in the absence of oxygen, and Alcaligenes appears to be a potential candidate for this use. Recent research is looking at the use of a polarized electrode made from carbon cloth, rather than oxygen, as the final electron acceptor in ammonia oxidation in stain HO-1. This would have the potential to reduce aeration in wastewater treatment plants while still efficiently removing nitrogen through the anaerobic denitrification process that is dominant in wastewater treatment (Pous et al. 2023).
References
Gao, X.Y., Xie, W., Liu, Y., Ma, L. and Liu, Z.P., 2022. Alcaligenes ammonioxydans HO-1 antagonizes Bacillus velezensis via hydroxylamine-triggered population response. Frontiers in Microbiology, 13.
Hendrie, M.S., Holding, A.J. and Shewan, J.M., 1974. Emended descriptions of the genus Alcaligenes and of Alcaligenes faecalis and proposal that the generic name Achromobacter be rejected: status of the named species of Alcaligenes and Achromobacter: request for an opinion. International Journal of Systematic and Evolutionary Microbiology, 24(4), pp.534-550.
Hou, T.T., Miao, L.L., Peng, J.S., Ma, L., Huang, Q., Liu, Y., Wu, M.R., Ai, G.M., Liu, S.J. and Liu, Z.P., 2022. Dirammox Is Widely Distributed and Dependently Evolved in Alcaligenes and Is Important to Nitrogen Cycle. Frontiers in Microbiology, 13.
Pous, N., Bañeras, L., Corvini, P.F.X., Liu, S.J. and Puig, S., 2023. Direct ammonium oxidation to nitrogen gas (Dirammox) in Alcaligenes strain HO-1: The electrode role. Environmental Science and Ecotechnology, 15, p.100253.
Wu, M.R., Hou, T.T., Liu, Y., Miao, L.L., Ai, G.M., Ma, L., Zhu, H.Z., Zhu, Y.X., Gao, X.Y., Herbold, C.W. and Wagner, M., 2021. Novel Alcaligenes ammonioxydans sp. nov. from wastewater treatment sludge oxidizes ammonia to N2 with a previously unknown pathway. Environmental Microbiology, 23(11), pp.6965-6980.
Author
Trevor Tubbs, a student of Dr. Hidetoshi Urakawa of Florida Gulf Coast University for Microbial Ecology EVR 4024C spring 2023.