Antibiotics and Antibiotic Resistance in Nature
Introduction
According to the Oxford Dictionary, the term Antibiotics encompass medicines (such as penicillin or its derivatives) that inhibit the growth of or destroys microorganisms. Antibiotics are naturally occurring substances that exhibit inhibitory properties towards microbial growth at high concentrations [1]. Today, most antibiotic drugs are semi-synthetic in that their natural chemical structures have been modified [1]
Origin of Antibiotics
Alexander Fleming [1] discovered penicillin, the first class of antibiotics, in the 19th century. He observed the inhibitory nature of an accidental fungal contamination on the growth of cultured bacteria. This discovery led to the isolation and cultivation of penicillin as a drug. A novel compound that heralded humanity into a new age of decreased mortality, disease spread and general improvements to quality of life [1]. Due to the initial widespread use, bacterial resistance to the penicillin compound became increasingly prevalent, prompting further research into alternatives. Thus spawning the creation of the first classes of semi-synthetic antibiotics: oxacillin [2], meticillin [3], cloxacilin [4] and dicloxacillin [5] [1]. Humanity’s increased endeavour in understanding penicillin’s beta-lactam structure led to the semi-synthesis of the second generation antibiotics: ampicillin and amoxicillin [1]. As the class of beta-lactam penicillin’s grew, other classes of antibiotic compounds with different mechanisms of action were also discovered (such as streptomycin) [1]. Spurred by emerging bacterial resistance to each antibiotic, more and more discoveries were made in the expansion and creation of different class types distinct in their modes of action, specificity and range; ultimately resulting in the repertoire of antibiotic compounds we have today [1].
Mechanism of Activity
The inhibitory characteristic of antibiotics in the clinical setting comes from the ability of the compounds to disrupt specific processes within growing bacterial cells, indirectly resulting in cell death. Each antibiotic class have characteristic functional groups that dictate how and which cellular process is inhibited.
Penicillin
The beta-lactam structure is iconic for all antibiotics of the penicillin family and is important in sequestering the penicillin binding protein (PBP) [6] involved in bacterial cell wall synthesis. PBP catalyzes the peptide cross linkage bonds critical in maintaining structural integrity of the bacterial peptidoglycan cell wall. With PBP sequestered by penicillin, the cross linkage bonds are not formed and the cell is lysed due to turgor pressure from the hypotonic environment [2].
Cephalosporin
Cephalosporin antibiotics have an additional six membered, heterocyclic, sulphur-containing ring attached to a beta-lactam structure. They are similar to penicillin’s in their mechanism of action [2].
Flouroquinolone
Unlike the penicillin and cephalosporin classes, Flouroquinolones are not based on natural products but are completely synthetic (initial starting compound is nalidixic acid). Flouroquinolones function to inhibit proper unwinding of bacterial DNA during replication thereby halting the process and resulting in cell death [2].
Tetracycline
Tetracyclines are four-membered ring structures with various functional groups attached produced by bacteria of the Streptomyces species. They disrupt protein synthesis by binding to the larger portion of the cellular ribosome, thus blocking the attachment of amino acid bound tRNA’s [2].
Macrolides
Macrolides, produced by Streptomyces erythraeus [7], have a complex structure characterized by a large lactone ring with two sugar molecules attached via glycoside bonds. Antibiotics in this class inhibit protein synthesis in a functionally similar way as tetracyclines [2].
Aminoglycosides
Aminoglycosides inhibit protein synthesis in a generally similar mechanism as tetracyclides and macrolides [3].
Role of Antibiotics in Nature
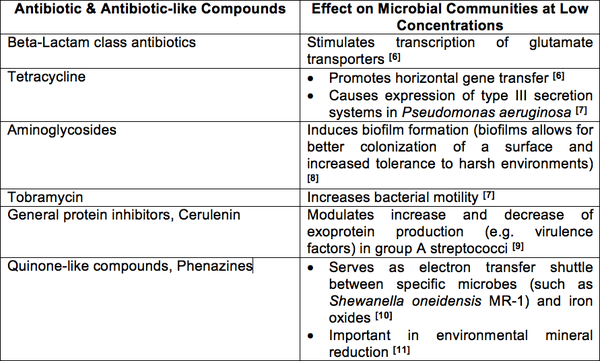
Based on clinical use of antibiotics, it may appear that these compounds play a similar role as microbial weapons in nature, yet this seems unlikely due to the fact that the concentrations used in the clinical setting are significantly higher than that produced in nature [4]. Based on experimental evidence, it makes more sense to see antibiotics as small, secreted molecules involved in cell-to-cell communication within microbial communities [5]. Studies have been conducted in which different antibiotics and antibiotic-like structures were administered to different bacterial species at levels below the compounds minimum inhibitory concentrations (MIC) [4]. What was observed is that sub-inhibitory concentrations of different antibiotics elicit certain phenotypic and even metabolic changes in specific bacterial species; changes that, in most cases, are beneficial to the target organism or community [4].
If antibiotic compounds play the role of effector molecules in their natural environments, then their mechanism of action could be seen as using their specific targets as signal relays [12]. For example, tetracyclines binding to the ribosome cause a breakdown in translation that ultimately lead to cell death at high concentrations; but at sub-inhibitory concentrations, the binding to the ribosomes causes a temporary stall in translation that leads to an mRNA build up thereby stabilizing mRNA transcript concentrations in target cells [4]. Therefore, the action of antibiotic is actually an interesting way of using ribosomes, DNA, RNA, carbohydrates (peptidoglycan) as signal effector receptors and transducers [4].
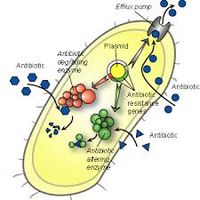
Role of Antibiotic Resistance in Nature
If antibiotics in the natural environment play such a different roles than that observed in clinical settings, then this also implies that antibiotic resistance in nature is not completely a defensive mechanism. Antibiotic resistance is a trait that is said to have existed before humanity’s use of antibiotics [13], with some species having the genes regardless of human intervention [12]. With signalling compounds being secreted from multiple sources in the soil microenvironment, It has been theorized that antibiotic resistance is a way for microbial communities to selectively regulate the response to these signalling molecules [14]; thus deciding which species biochemical pathways are affected and which are not [12]. This system of regulation has been observed in the characteristic differentiation of antibiotic resistant mutants compared to their wild type parents when both are exposed to similar antibiotics [6].
Relevancy to Humans
With antibiotics being such a critical therapeutic factor in human disease control, resistance to these compounds is a critical issue. Ironically, it is human misuse and overuse of antibiotics that is promoting the success of resistance genes in the environment. By introducing such high doses of antibiotics in the environment through agricultural [12] use and increasing the prevalence of resistance genes through the misuse of antibiotics, the microbial population dynamic in nature has been affected [13]. With antibiotic communication being key to phenotypic characteristics and biochemical pathways, an increase in resistance could mean drastic changes in metabolic and phenotypic traits of microbial communities and ecosystems [5]. Another consequence of increased resistance genes due to human activity is the possibility for horizontal gene transfer of said genes to pathogenic bacteria [12]. Losing our weapons against pathogens could mean humanity’s return to the state it once was before the discovery of antibiotics, where infections like syphilis mean the death of the patient [1].
Current Research
A more optimistic analysis is the possibility that there exists millions more signalling molecules yet to be characterized with some possibly possessing antibiotic activity [14]. UBC’s Julian Davies is currently looking at profiles of soil microbiota in an effort to discover new and different compounds that could potentially lead to new classes of antibiotics against pathogens resistant to the current compounds [14].
References
[1] Zaffiri, Lorenzo, Jared Gardner, and Luis Toledo-Pereyra. "History of Antibiotics. From Salvarsan to Cephalosporins." Journal of Investigative Surgery 25.2 (2012): 67-77. Print.
[2] Sköld, Ola. Antibiotics and Antibiotic Resistance. Hoboken, NJ: Wiley, 2011. Print.
[3] Avent, M. L., B. A. Rogers, A. C. Cheng, and D. L. Paterson. "Current Use of Aminoglycosides: Indications, Pharmacokinetics and Monitoring for Toxicity." Internal Medicine Journal 41 (2011): 441-49. Print.
[4] Fajardo, A., and J. Martinez. "Antibiotics as Signals That Trigger Specific Bacterial Responses." Current Opinion in Microbiology 11.2 (2008): 161-67. Print.
[5] Martinez, J. L. "Antibiotics and Antibiotic Resistance Genes in Natural Environments." Science 321.5887 (2008): 365-67. Print.
[6] Yim, Grace, Helena Huimi Wang, and Julian Davies. "Antibiotics as Signalling Molecules." Philosophical Transactions of the Royal Society B: Biological Sciences 362.1483 (2007): 1195-200. Print.
[7] Linares, J. F., I. Gustafsson, F. Baquero, and J. L. Martinez. "Antibiotics as Intermicrobial Signaling Agents Instead of Weapons." Proceedings of the National Academy of Sciences 103.51 (2006): 19484-9489. Print.
[8] Hoffman, Lucas R., David A. D'Argenio, Michael J. MacCoss, Zhaoying Zhang, Roger A. Jones, and Samuel I. Miller. "Aminoglycoside Antibiotics Induce Bacterial Biofilm Formation." Nature 436.7054 (2005): 1171-175. Print.
[9] Tanaka, M., T. Hasegawa, A. Okamoto, K. Torii, and M. Ohta. "Effect of Antibiotics on Group A Streptococcus Exoprotein Production Analyzed by Two-Dimensional Gel Electrophoresis." Antimicrobial Agents and Chemotherapy 49.1 (2004): 88-96. Print.
[10] Lies, D. P., M. E. Hernandez, A. Kappler, R. E. Mielke, J. A. Gralnick, and D. K. Newman. "Shewanella Oneidensis MR-1 Uses Overlapping Pathways for Iron Reduction at a Distance and by Direct Contact under Conditions Relevant for Biofilms." Applied and Environmental Microbiology 71.8 (2005): 4414-426. Print.
[11] Hernandez, M. E., A. Kappler, and D. K. Newman. "Phenazines and Other Redox-Active Antibiotics Promote Microbial Mineral Reduction." Applied and Environmental Microbiology 70.2 (2004): 921-28. Print.
[12] Allen, Heather K., Justin Donato, Helena Huimi Wang, Karen A. Cloud- Hansen, Julian Davies, and Jo Handelsman. "Call of the Wild: Antibiotic Resistance Genes in Natural Environments." Nature Reviews Microbiology 8.4 (2010): 251-59. Print.
[13] Martinez, J. L. "The Role of Natural Environments in the Evolution of Resistance Traits in Pathogenic Bacteria." Proceedings of the Royal Society B: Biological Sciences 276.1667 (2009): 2521-530. Print.
[14] Mlot, C. "Antibiotics in Nature: Beyond Biological Warfare." Science 324.5935 (2009): 1637-639. Print.