Arenaviridae
A Viral Biorealm page on the family Arenaviridae
Baltimore Classification
Higher order taxa
Virus; ssRNA negative-strand viruses; Arenaviridae
Genera
Arenavirus
Description and Significance
Arenaviruses are generally associated with rodent-transmitted diseases in humans. Each virus is usually associated with a particular rodent host species in which it is maintained. Arenavirus infections are relatively common in humans in some areas of the world and can cause severe illnesses.
The virus gets its name from the Latin word 'arena' which means 'sandy'. This is because they acquire grainy particles, ribosomes, from their host cells.
Lymphocytic Choriomeningitis virus (LSMV) was the first arenavirus to be isolated in 1933 during a study of an epidemic of St. Louis Encephalitis.
These viruses are zoonotic and are found in animals in nature. The rodent hosts of arenaviruses are chronically infected with the viruses but the viruses donot seem to cause any illness to the rodents. Humans can get infected with the arenaviruses when an individual comes into contact with the excretions or materials contaminated with the excretions of infected rodents. Aerosol transmission is also possible: inhalation of tiny particles soiled with rodent urine or salive can also cause infection. Secondary person-to-person and nosocomical transimission are also sometimes possible, usually associated with direct contact with the blood or other excretions containing virus particles of infected individuals.
Genome Structure
The virus has a segmented genome that consists of linear, negative-sense to ambisense, single-stranded RNA. The genome is transcriptional inactive. Minor species of non-genomic nucleic acid are also found in virions. The encapsidated nucleic acid is mainly of genomic origin, but virions may also contain subgenomic RNA and nucleic acid of host origin including three molecules of host rRNA. RNA segments are not homologous. The complete genome is about 10000-11000 nucleotides long. The RNA-L is sequenced, and the complete sequence is 7400 nucleotides long while RNA-S is about 3400 nucleotides long. The genome has a guanine+cytosine content of 40-45%. Although not covalently closed, the genome has a circular virus coded terminal protein. Nucleotide sequences at the 3'-terminus are largely complimentary to simial regions on the 5' end. The 5'-end of the genome does not have a cap. The 3'-terminus has conserved nucleotide sequences; in all segments and species of the same genus; sequence has 19-30 nucleotides in length; in S RNA. The intergenic region has a S hairpin configuration. The multipartite genome is encapsidated, each segment in a separate nucleocapsid, and the nucleocapsids are surrounded by one envelope. Each virion contains multiple copies of the genome; often segments of the genome in non-equimolar proportions due to the frequent packaging of S RNA. (source: ICTV dB Descriptions)
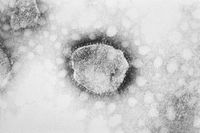
Virion Structure of an Arenovirus
The virions of arenavirus consist of an envelope and an enveloped nucleocapsid. Virions are spherical to plemorphic. The envelope has surface projections and surrounds two nucleocapsids (probably). The surface projections are distinctive club-shaped peplomers that are spaced widely apart and cover the surface evenly. They are embedded in a lipid bilayer that comprises of surface glycoproteins (GP). Surface projections are composed of one type of protein and they are 8-10 nm long. Host ribosmes are seen inside the envelope in varying numbers. The capsid is elongated with helical symmetry. Virions consist of two nucleocapsids. The nucleocapsid is filamentous and formes a closed circle. It appears like a 'string of beads' and varies in length from 1000-1300nm for L segment, 450-640nm for S segment and is 3-4nm wide. The nucleocapsid contains a polymerase complex and a nucleoprotein complex. The nucleocapsids are isolated, free of contaminating host ribosomes organized in closed circles and display a linear array of nucleosomal subunits. (source: ICTV dB Descriptions)
Arenavirus enters the host cell by targetting activated monocytes. The alpha-dystroglycan molecule is surface receptor for both Lassa Fever Virus and Lymphocoriomeningitis virus. There is evidence for both pH mediated fusion and direct plasma membrane fusion. Once in the cell, arenavirus makes nucleoprotein and more transcriptase. Arenavirus steals cap from cellular RNA and replication occurs. During the replication, there are two rounds of transcription to cope with the ambisense coding stratergy. One transcription takes place before the formation of a 'reverse-sense' RNA intermediate while another transcription takes place after the formation. An open end reading frame at 3' end of vRNA is transcribed and a cap is stolen from the host. The mRNA is is translated into the N protein and the genome is replicated. Then an open reading frame at the 3' end of vcRNA can be transcribed and a cap is stolen from the host. The mRNA is then translated to NSs protein. (source: SOM 208 MICROBIOLOGY SYLLABUS)
Viral Ecology & Pathology
Cases of LCM and mild LCMV infections have been reported in Europe, the Americas, Australia, and Japan, and may occur wherever infected rodent hosts of the virus are found. There has been a problem of underreporting of the disease which has made it difficult to determine incidence rates or estimates of prevalence by geographic region. Several serologic studies conducted in urban areas have shown that the prevalence of LCMV infection among humans ranges from 2-5%.
Arenavirus produces chronic infections in its natural hosts, the rodents. The virus could also establish a persistent infection in cell lines. The virus infects man very rarely, causing mild diseases. (source: Microbiology and Immunology Online: University of South Carolina)
References
Microbiology and Immunology Online: University of South Carolina
