Azotobacter chroococcum
Classification
Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae
Species
|
NCBI: [1] |
Azotobacter chroococcum
Description and Significance
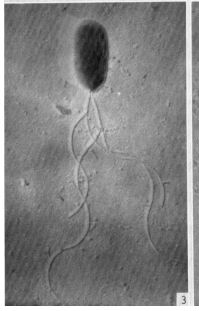
Azotobacter chroococcum colonies are characterized by a creamy color, circular shape, and smooth convex type. Single bacterium are characterized by a rod shape and are motile by way of flagella (Talabani et al. 2019). This bacteria inhabits neutral and slightly alkaline soils, with an optimal pH range of 7.1-9.0. The strongest influence of Azotobacter chroococcum abundance in soil is the total nitrogen and organic carbon contents. The strong correlation of Azotobacter chroococcum abundance with total nitrogen content is most likely due to its nitrogen fixation ability (Lenart 2012). This bacteria is important not only to the nitrogen cycle, but also to agricultural sustainability and soil remediation.
Genome Structure
Only one strain of the bacterial species Azotobacter chroococcum has been sequenced in its entirety. The genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412)(Ac-8003) consists of a singular, circular chromosome and six plasmids pAcX50a, b, c, d, e, f totaling 5,192,291 bp. The chromosome contains 4,591,803 bp while the six plasmids contain 10,435 bp, 13,852 bp, 62,783 bp, 69,713 bp, 132,724 bp, and 311,724 bp respectively. The chromosome has a G+C content of 66.27% while the six plasmids have a G+C content of 58.1%, 55.3%, 56.7%, 59.2%, 61.9%, and 62.6% respectively. The genome also encodes 4628 predicted protein-encoding genes and 258 transposase (Robson et al. 2015).
Cell Structure, Metabolism and Life Cycle
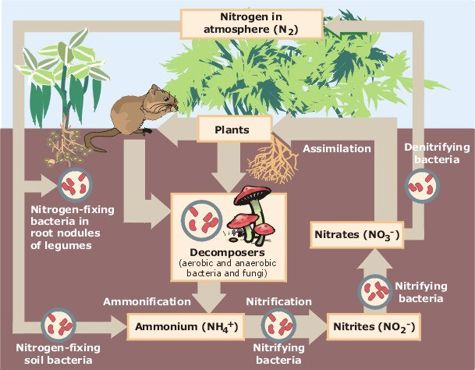
Azotobacter chroococcum is a free-living, obligately-aerobic, gram negative bacterium that has the ability for atmospheric nitrogen fixation (Robson et al. 2015, Talabani et al. 2019). These bacteria are motile by way of polar flagella, and move towards root exudates by chemotaxis (Kumar et al. 2007). Azotobacter chroococcum is a heterotroph, and obtains carbon through fructose, galactose, glucose, trehalose, starch, glycogen, ethanol, butan-1-ol, pentan-1-ol, mannitol, sorbitol, myo-inositol, and caproate (Robson et al. 2015). Through the process of nitrogen fixation, Azotobacter chroococcum produces ammonia which can readily be taken up by plant roots. This bacterium also produces polymers such as alginates, levans, cellulose, capsule lipopolysaccharides, and antimicrobials for protection and adhesion. Siderophores are produced for nutrient acquisition in times of low nutrient availability. Melanin is produced to minimize oxidative stress and carotenes are produced as antioxidants (Robson et al. 2015).
Ecology and Pathogenesis
Azotobacter chroococcum is a free-living bacterium in the soil. This bacteria occurs mostly in neutral and slightly alkaline soils, with an optimum pH range of 7.1-9.0 (Lenart 2012). Even though it is free living, this bacteria experiences a mutualistic relationship with plants. Azotobacter chroococcum provides plants with fixed nitrogen compounds and protection from soil-born pathogens while the plants provide it with complex carbohydrates in the form of root exudates (Kumar et al. 2007). Due to this mutualistic relationship, Azotobacter chroococcum plays a major role in the global nitrogen cycle. Atmospheric nitrogen fixation occurs through use of molybdenum and vanadium nitrogenase systems to produce ammonia available in the soil (Robson et al. 2015).
References
Bisset KA, Hale CMF. 1953. The Cytology and Life-cycle of Azotobacter chroococcum. Journal of General Microbiology. [accessed 2022 December 12;8(3):442-448. doi:10.1099/00221287-8-3-442.]
Kumar R, Bhatia R, Kukreja K, Behl RK, Dudeja SS, Narula N. 2007. Establishment of Azotobacter on plant roots: chemotactic response, development and analysis of root exudates of cotton (Gossypium hirsutum L.) and wheat (Triticum aestivum L.). Journal of Basic Microbiology. [accessed 2022 November 21;47(5):436-439. doi:10.1002/jobm.200610285.]
Robson RL, Jones R, Robson RM, Schwartz A, Richardson TH. 2015. Azotobacter Genomes: The Genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412). PLOS ONE. [accessed 2022 November 20;10(6):e0127997. doi:10.1371/journal.pone.0127997.]
Talabani SK, Fattah OA, Khider AK. 2019. CLASSICAL AND MOLECULAR APPROACHES FOR IDENTIFICATION OF RHIZOBIUM LEGUMINOSARIUM, AZOTOBACTER CHROOCOCCUM AND BACILLUS MEGATERIUM. APPLIED ECOLOGY AND ENVIRONMENTAL RESEARCH. [accessed 2022 November 21;17(5):12491-12506. doi:10.15666/aeer/1705_1249112506.]
Author
Page authored by Isabella Ickes, student of Prof. Bradley Tolar at UNC Wilmington.