Bacteroides coprosuis

Introduction to Bacteroides
"Bacteroides species are significant clinical pathogens and are found in most anaerobic infections, with an associated mortality of more than 19%. The bacteria maintain a complex and generally beneficial relationship with the host when retained in the gut, but once they escape this environment they can cause significant pathology, including bacteremia and abscess formation in multiple body sites".
Classification
Higher order taxa
Domain: Bacteria;
Phylum:Bacteroidales;
Class:Bacteroidia;
Order:Bacteroidales;
Family: Bacteroidaceae;
Genus:Bacteroides
Species
Bacteroides coprosuis
Description and significance
Coprosuis belongs to the genus Bacteroides. Members of the genus Bacteroides in general protect animal guts from pathogenic microorganisms and they also break down complex molecules such as polysaccharides. Bacteroides coprosuis was isolated from a manure storage pit of a swine facility;however, it is not found in an animal host.Therefore, the exact habitat remains unknown. The type strain is PC 139.
Cell structure/growing environment
Bacteroides coprosuis cells are Gram-negative, anaerobic, non-motile and non-spore-forming. They are rod-shaped organisms. Typical cells are 0·8–3·0 μm by 0·5–1·5 μm in size. The optimum temperature for Bacteroides coprosuis is 37 °C, but they can still grown within 25 to 37 °C range. It will not grow at 42 °C or higher. When growing in lab,Bacteroides coprosuis was able to grow on media that contain glucose, maltose and chondroitin sulfate but, no growth was observed on arabinogalactan, arabinose, cellobiose, corn-fibre xylan, corn-spelt xylan, fructose or xylose. As a result, its fermentation products from glucose are acetic acid (8.0-15.0 mM), succinic acid (7.5-10.0 mM) and propionic acid (4.0-22.0 mM).
Antibiotic characteristics
According to a research study by Wexier, “species of the genus Bacteroides have the most antibiotic resistance mechanisms and the highest resistance rats of all anaerobic pathogens.Clinically, Bacteroides species have exhibited increasing resistance to many antibiotics, including cefoxitin, clindamycin, metronidazole, carbapenems, and fluoroquinolones ”. Bacteroides Coprosuis itself shows resistant to ampicillin (100 µg/ml), cefoxitin (20 µg/ml), erythromycin (10 µg/ml), gentamicin (200 µg/ml) and tetracycline (3 µg/ml).
"Bacteroides coprosuis shows activity for alkaline and acid phosphatase, α-fucosidase, β-galactosidases, α- and β-glucosidases, N-acetyl-β-glucosaminidase, chymotrypsin, esterase C4, ester lipase C8, lipase C14, cystine arylamidase, leucyl glycine arylamidase, alanine arylamidase, arginine arylamidase and glutamyl glutamic acid arylamidase. No activity was detected for urease, catalase, oxidase, trypsin, arginine dihydrolase, β-galactosidase 6-phosphate, β-glucuronidase, α-arabinosidase, α-mannosidase and glutamic acid, glycine, histidine, leucine, phenylalanine, proline, pyroglutamic acid, serine, tyrosine and valine arylamidase".
Similar microbe from the same location
PC111 was also isolated from the same location (a manure storage pit from a swine facility near Peoria, IL, USA). It was also anaerobic, non-spore-forming, non-motile and gram negative rod.Strain PC 139 and PC111 share some interest similarities. Researchers discovered that both strains were genetically highly related to each other. They displayed 99.7% 16S rRNA gene sequence similarity. PC111 also shows resistant to ampicillin (100 µg/ml), cefoxitin (20 µg/ml), erythromycin (10 µg/ml), gentamicin (200 µg/ml) and tetracycline (3 µg/ml).
Genome
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain CCUG 50528T is AF319778.
The DNA G+C content of the PC139 strain is 35.0 mol% (It 36.4 mol% in Terence’s research study). The genome consists of a 2,991,798 bp long circular chromosome. Out of the 2,539 genes predicted, 2,461 were protein-coding genes, and 78 RNAs; 68 pseudogenes were identified as well. The majority of the protein-coding genes (66.4%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins (Land, 2011).
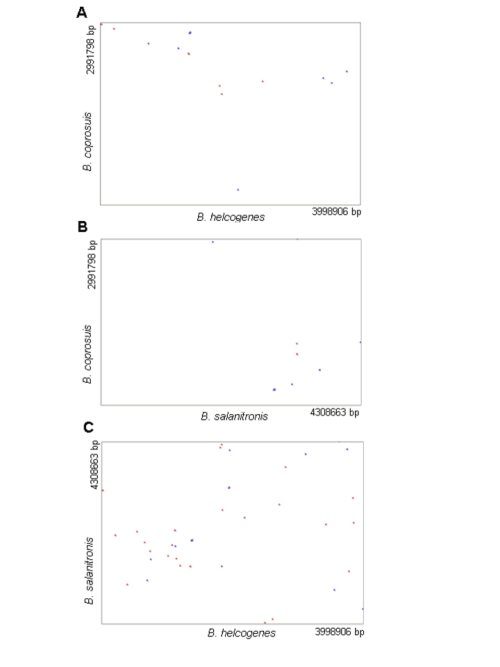
Bacteroides coprosuis VS Bacteroides helcogenes VS Bacteroides salanitronis
Genome sequences were compared among Bacteroides coprosuis, Bacteroides helcogenes and Bacteroides salanitronis. Researchers concluded that the genome size of Bacteroides coprosuis (3 Mb) is significantly smaller than Bacteroides helcogenes (4 Mb) and Bacteroides salanitronis (4.3 Mb). Also the G+C content of the Bacteroides coprosuis is much lower than the G+C content of Bacteroides helcogenes and Bacteroides salanitronis. Overall,Bacteroides salanitronis and Bacteroides helcogenes share more genes (543 genes) that are not present in Bacteroides coprosuis .
References
Edited by (LiWen Chen), student of Rachel Larsen at the University of Southern Maine