CCR5, Delta-32 Mutation and the HIV Infection Pathway
By Jon Funder Hansen
Introduction
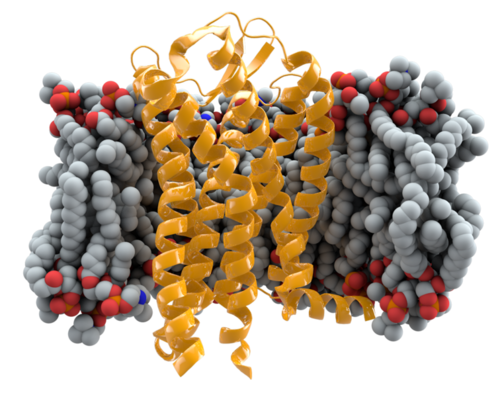
CCR5 (C-C chemokine receptor type 5) is surface protein located on the plasma membrane of white blood cells. CCR5 acts as a cytokine receptor and is primarily expressed on cells involved in the immune response, such as T-cells and macrophages. Individuals with mutations in one or both of the CCR5 alleles exhibit resistance to the Human Immunodeficiency Virus type 1 (HIV-1), the most common strain of the virus, as HIV-1 uses CCR5 as a co-receptor of infection.
In 1995 it was observed that HIV-1 infection of T-lymphocytes can be inhibited by human chemokines.[10] The following year it was discovered that HIV-1 infection was mediated by a G-protein-coupled receptor with seven transmembrane segments and that two of these receptors are CCR3 and CCR5[11][12][13][14]. At around the same time, a polymorphic expression of the CCR5 gene that leads to resistance to HIV-1 infection was identified[15]. This led to the discovery that one or more human chemokine receptors, such as CCR5, are necessary as co-receptors of HIV-1 infection[12].
The Structure and Function of CCR5
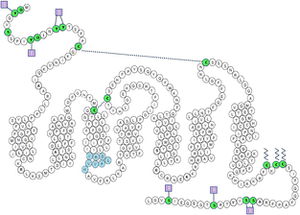
CCR5 is comprised of 352 amino acids and has a molecular weight of 40.6 kDa[16]. The protein contain seven trans-membrane domains, an amino terminal, a carboxyl tail terminal and three intracellular loops. The important regions for a functional response of the receptor, chemokine ligand binding and HIV-1 coreceptor activity include the specific motifs of hydrophobic or charged regions, the conserved residues and several post translational modifications[18].
Chemokines are proteins that are involved in the direction of immune cell migration in the body. There are over fifty human chemokines that all work by binding to G-protein-coupled chemokine receptors, such as CCR5, on the surface of cells. Chemokines are grouped into two major families, inflammatory and homeostatic. CCR5 is one of the eighteen human chemokine receptors that have been identified so far. CCR5 regulates both the trafficking and effector molecule production of T-cells, immature dendritic cells and macrophages.[1]
CCR5 is one of three high affinity receptors of CCL5 (along with CCR1 and CCR3), which is an inflammatory chemokine that acts as one of the key regulators of T-cell inflammatory migration, directing the T-cells towards sites of infection.[2] Research suggest that CCL5:CCR5 Axis mediated signaling could serve an important role in mucosal Trypanosoma cruzi protection as well as B-cell activation. The CCR5-CCL5 Axis may be critical for the optimal control of T. cruzi replication.[3] It has been shown that CCR5-CCL5 interactions provide antiapoptotic signals that increase macrophage survival during mouse parainfluenza and human influenza infections in mice. The lack of the CCL5 chemokine in infected mice cause immunodeficiency, leading to excessive airway inflammation, delayed viral clearance and respiratory death. CCL5 protection requires the activation of the CCR5 receptor to stop cell to cell infections by macrophages.[4]
The Role of CCR5 in HIV Infection
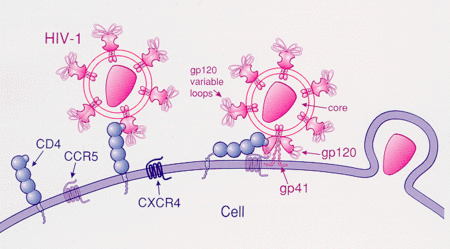
The replication cycle of HIV first starts by the virus being transmitted from one host to another. This may occur through the transmission of semen, vaginal fluid, rectal fluids, blood and breast milk, or alternatively from one cell to another inside the body. Like all viruses, HIV depend on host cells to reproduce. To infect organisms, HIV first needs to recognize and attach to certain receptor molecules on the surface of specific cells. The leukocyte glycoprotein receptor (CD4) is a surface protein found on CD4 T lymphocytes that acts as the primary receptor for HIV. CD4’s regular function is to connect T lymphocytes with antigen presenting cells, which activates the production of antibodies by B-cells.[9] Spike proteins, consisting of three gp120s (envelope glycoprotein gp120) and gp41s, mediates the attachment and membrane fusion between the host cell and the virus.[9]
In HIV-1 infections, the attachment begins with an interaction between the gp120 and the CD4 protein on the surface of the cell. The binding of CD4 exposes the third variable region (V3) loop of the gp120 which allows it to bind with a chemokine receptor, CCR5 or CXCR4. Once the V3 loop is exposed, it interacts with both the ECL2 and the N-terminus of the CCR5, which causes the spike protein to release gp41. Two structures in the gp41 called HR1 and HR2 rearrange and folds into a six helical bundle, that exposes the gp41 fusion peptides.[8] The peptides are extended into the host cell membrane, where they facilitate the fusion between the virus and cell membrane.[5]
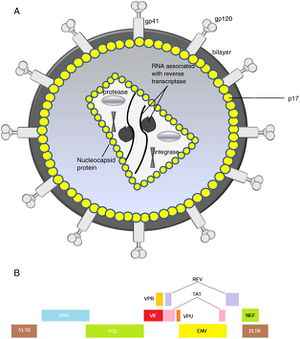
HIV is an RNA retrovirus, which means that HIV needs to use reverse transcriptase, usually bound to the RNA as a part of the virion, to copy its RNA genome into DNA. The HIV virion carries two (+) RNA strands of its genome, each with an reverse transcriptase attached. Once the RNA has been transcribed into DNA, the DNA is transported across the nuclear membrane where it is integrated into the host genome through the help of the enzyme integrase. The DNA embedded in the host genome is then transcribed into new viral RNA as both genomic RNA and to be translated into viral proteins. Some of these proteins condense into a capsid that encapsulates the genomic RNA along with other viral proteins, which then move to the cellular membrane and forms a new HIV virion.[9]
CCR5 Mutation
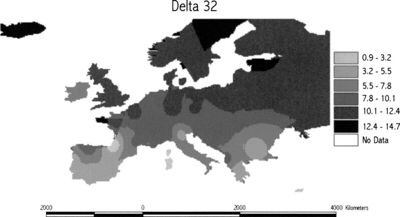
CCR5-Δ32 Mutation
CCR5-Δ32 is a 32-base pair deletion mutation of the CCR5 gene in the region that corresponds to the second extracellular loop of the CCR5 receptor.The mutation was first discovered in 1996 as a mutation that protects cells from being infected by HIV. Genetic analysis of the gene revealed that the mutation consists of a 32 basepair deletion from nucleotide 794 to 825[17]. The mutation also involves a frameshift mutation which confer the inclusion of seven amino acids from amino acid number 174 followed by a stop codon[16]. The regular wild type CCR5 has a total length of 352 amino acids, the mutation, in comparison, only expresses 215[18]. This renders the CCR5 receptor non-functional, which results in the reduced ability of the HIV-1 virus to enter the cell.
CCR5-Δ32 is common among Caucasian populations, typically around 10%, and is found at lower frequencies in India and the Middle East. Haplotype analysis has been used to indicate that the mutation originated approximately 700 years ago (with a range of 275-1875 years) in a single individual in northeastern Europe. A gradient cline in the allele frequencies of the mutation has been found from northern to southern Europe, suggesting that the mutation increased the survival of Caucasian populations against a strong selective factor such as a pathogen.[6] It has been hypothesized that the mutation could have conferred resistance against the Black Death and thus become favored by natural selection. However, further research has shown that the gene mutation does not provide protection against Yersinia pestis infections. [7] The current hypothesizes suggest that the mutation may have been naturally favored because it could have conferred protection against smallpox or an hemorrhagic diseases similar to the Ebola-like virus.[7]
Much speculation and research has gone into investigating the cause of the rapid spread of the mutation across Europe and to other parts of the world. Some have suggested that the mutation has been disseminated across Europe and beyond by Scandinavian Vikings during the Viking Age.[6] Others have suggested that the distribution of allele frequency may not have been caused by gene spreading, but rather by negative selection pressures resulting in the spread of pathogens during the expansion of the Roman Empire.[7]
Other Mutations
The strong resistance to HIV-1 infection among individuals that are homozygous for the delta-32 allele has driven researchers to search for similar mutations in different human populations. Sixteen other naturally occurring CCR5 mutations have been found in populations throughout the world. Each of these mutations are generally restricted within specific populations with an allele frequency ranging from one to five percent. Out of these sixteen, six have been shown to express major alterations in regards to their functional response to chemokines, ligand binding affinities or inability to confer receptor activation. The relatively high frequency of nonfunctional CCR5 alleles among various populations reinforce the suggestion that a strong selective pressure has been favoring these mutations.Out of the six mutations that express major alterations in the functionality of the protein, only C101X have been found to be completely unable to mediate HIV-1 infection[19].
The C101X mutation cause a premature termination of translation and has a relatively high allele frequency in central Africa. The mutation was discovered in a homosexual, HIV negative man who had frequently been exposed to HIV. The mutation results in a substitution at amino acid 101, as cysteine is replaced by a stop codon. As a result of the premature termination of translation, the protein produced contain just two trans-membrane domains.
In vitro studies have indicated that the protein produced is misfolded, not expressed on the surface of the cell, and therefore does not function as a co-receptor of HIV-1 infection[19].
Therapeutic Applications of CCR5 Against HIV Infection

Individuals that are homozygous for the Delta-32 mutation, and therefore do not express the CCR5 receptor, appears to have no disadvantages in terms of health as a result of the lack of CCR5 expression. However, whether or not the Delta-32 mutation impart a protective or negative effect against other diseases is still a controversial topic[16].
The primary application of CCR5 against HIV infection lies in the potential for using receptor antagonists to bind to CCR5 and prevent infection. Clinical trials have been performed on a number of CCR5 antagonist, only one of which, maraviroc, was approved for treating HIV infected individuals in 2007. In addition to using CCR5 antagonists for treatment of infected patients, they have many different potential uses, such as preventing HIV transmission and preventing the rejection of transplanted organs[20].
In 2008, Timothy Ray Brown got what scientist call a sterilizing cure. Brown was diagnosed with HIV in 1995 and later, in 2006, was also diagnosed with acute myeloid leukemia. In 2007 and 2008 Mr. Brown received two stem cell transplants from a single donor with homozygous alleles for the delta-32 mutation. After the initial transplant, Mr Brown stopped his antretroviral medication treatment, which three months later resulted in the plummeting of his HIV infection levels and increased the number of CD4 T cells in his blood. To this day, Mr. Brown is not using antiretroviral drugs and is considered cured by the scientific community[21]. This has lead other scientists to propose a similar mode of action though the use of genetically engineered autologous cells to treat HIV by using gene therapy[16].
Conclusion
The C-C chemokine receptor type 5 truly is a fascinating surface protein that have played a fundamental role in the lives of millions of people. The discovery of mutations in the CCR5 protein have shown and continue to show promising possibilities for the development of new treatments and cures against a virus and an illness that has taken the lives of 39 million individuals[22].
References
1. Oppermann, M. (2004). Chemokine receptor CCR5: Insights into structure, function, and regulation. Cellular Signalling, 16(11), 1201-1210. http://www.sciencedirect.com/science/article/pii/S0898656804000786
2. Tamamis, P., & Floudas, C. A. (2014). Elucidating a key anti-HIV-1 and cancer-associated axis: The structure of CCL5 (rantes) in complex with CCR5. Sci.Rep., 4. http://www.nature.com/srep/2014/140626/srep05447/full/srep05447.html
3. Sullivan, N. L., Eickhoff, C. S., Zhang, X., Giddings, O. K., Lane, T. E., & Hoft, D. F. (2011). Importance of the CCR5–CCL5 axis for mucosal trypanosoma cruzi protection and B cell activation. The Journal of Immunology, 187(3), 1358-1368. http://www.jimmunol.org/content/187/3/1358.long
4. Tyner, J. W., Uchida, O., Kajiwara, N., Kim, E. Y., Patel, A. C., O'Sullivan, M.,P., et al. (2005). CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nature Medicine, 11(11), 1180-1187.http://www.nature.com/nm/journal/v11/n11/full/nm1303.html
5. Tamamis, P., & Floudas, C. A. (2014). Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. Plos One, 9(4), e95767. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3999033/
6. Lucotte, G., & Dieterlen, F. (2003). More about the viking hypothesis of origin of the Δ32 mutation in the CCR5 gene conferring resistance to HIV-1 infection. Infection, Genetics and Evolution, 3(4), 293-295. http://www.sciencedirect.com/science/article/pii/S1567134803000960
7. Faure, E., & Royer-Carenzi, M. (2008). Is the european spatial distribution of the HIV-1-resistant CCR5-Δ32 allele formed by a breakdown of the pathocenosis due to the historical roman expansion? Infection, Genetics and Evolution, 8(6), 864-874. http://www.sciencedirect.com/science/article/pii/S1567134808001524
8. Tsibris, A. (2007). Update on CCR5 inhibitors: Scientific rationale, clinical evidence, and anticipated uses. The PRN Notebook, the Physicians’ Research Network, 12. http://www.prn.org/index.php/management/article/ccr5_inhibitors_hiv_86
9. Slonczewski, J., L., & Foster, J., W. (2014). In Twichell B. (Ed.), Microbiology an evolving science (3rd ed.). New York: W. W. Norton and Company.
10. Cocchi, F., DeVico, A. L., Garzino-Demo, A., Arya, S. K., Gallo, R. C., & Lusso, P. (1995). Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science (New York, N.Y.), 270(5243), 1811-1815.
11. Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., et al. (1996). Identification of a major co-receptor for primary isolates of HIV-1. Nature, 381(6584), 661-666.
12. Feng, Y., Broder, C. C., Kennedy, P. E., & Berger, E. A. (1996). HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science (New York, N.Y.), 272(5263), 872-877
13. Choe, H., Farzan, M., Sun, Y., Sullivan, N., Rollins, B., Ponath, P. D., et al. (1996). The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell, 85(7), 1135-1148.
14. Alkhatib, G., Combadiere, C., Broder, C. C., Feng, Y., Kennedy, P. E., Murphy, P. M., et al. (1996). CC CKR5: A RANTES, MIP-1a, MIP-1ß receptor as a fusion cofactor for macrophage-tropic HIV-1. Science, 272(5270), 1955-1958.
15. Liu, R., Paxton, W. A., Choe, S., Ceradini, D., Martin, S. R., Horuk, R., et al. (1996). Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell, 86(3), 367-377.
16. Barmania, F., & Pepper, M. S. (2013). C-C chemokine receptor type five (CCR5): An emerging target for the control of HIV infection. Applied & Translational Genomics, 2(0), 3-16. http://www.sciencedirect.com/science/article/pii/S2212066113000082#bb0510
17. Liu, R., Paxton, W. A., Choe, S., Ceradini, D., Martin, S. R., Horuk, R., et al. (1996). Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell, 86(3), 367-377.
18. Samson, M., Labbe, O., Mollereau, C., Vassart, G., & Parmentier, M. (1996). Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry, 35(11), 3362-3367.
19. Blanpain, C., Lee, B., Tackoen, M., Puffer, B., Boom, A., Libert, F., et al. (2000). Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood, 96(5), 1638-1645. http://www.ncbi.nlm.nih.gov/pubmed/10961858
20. Gilliam, B., Riedel, D., & Redfield, R. (2010). Clinical use of CCR5 inhibitors in HIV and beyond. Journal of Translational Medicine, 9, S9. http://www.translational-medicine.com/content/9/S1/S9
21. Hütter, G., Nowak, D., Mossner, M., Ganepola, S., Müßig, A., Allers, K., et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med, 360(7), 692-698. http://www.nejm.org/doi/full/10.1056/NEJMoa0802905#t=article
22. http://www.who.int/gho/hiv/en/
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
