Candida Krusei
1. Classification
a. Higher order taxa
Domain: Eukarya
Kingdom: Fungi
Phylum: Ascomycota
Class: Saccharomycetes
Order: Saccharomycetales
Family: Saccharomycetaceae
Genus: Candida
Species: Krusei
[1]
2. Description and significance
Candida krusei is a yeast species that does not produce spores that is usually found growing as a single cell or as pseudohyphae [1]. In the single cell form, the cells of C. krusei are round or ovoid and can be easily separated while in the pseudohyphae form the cells are more elongated and attached to neighboring cells [2]. C. krusei is considered a relatively uncommon, emerging opportunistic human pathogen [3] that infects mainly immunocompromised patients, especially those that have some form of leukemia or other deficiencies in white blood cells [4]. C. krusei is gaining recognition in the world today since it is a multidrug resistant pathogen due to its ability to rapidly adapt to antifungal treatments and its complex susceptibility profile [5]. C. krusei is most often found in the human microbiome in immunocompromised patients [4], in some foods, and it is used extensively in chocolate production from West African cocoa beans.
Before the 1960s, C. krusei was regarded as a species with low pathogenicity and virulence in humans compared to other species in the genus Candida [4]; however, since 1960, C. krusei infections have increased alarmingly. C. krusei is inherently resistant to the triazole antifungal, fluconazole which allows it to persist and cause infection in humans being treated with antifungals where other fungi cannot survive [6]. Of particular note is the high presence of C. krusei in the human microbiome in immunocompromised patients due to selection by prophylactic oral rinse therapy [2]. The increased use of fluconazole and other traizole antifungals to treat and quell fungal infections has thus led to a marked increase in infections by C. krusei , making it increasingly relevant in the world today [1]. However, although much is now known about C. krusei because of its status as an emerging pathogen, its genome has yet to be characterized and sequenced [7]. Additionally, a point of contention in the scientific community is whether or not C. krusei should be re-classified into a genus of its own since srRNA analysis and analysis of various ubiquinone systems in related species reveals C. krusei diverged much earlier than other Candida species [3].
3. Genome structure
C. krusei has eight chromosomes, in contrast to most members of the genus Candida, such as C. albicans, which has sixteen [1]. This difference, in addition to differences in cell structure and cell wall structure as compared to other Candida species, form the basis of the argument that C. krusei should be re-classified into a different genus [3]. While it is known how many chromosomes C. krusei has, the genome of C. krusei has yet to be sequenced [8], so the genome has not been fully characterized. However, increased infection rates of Candida species other than C. albicans [7] have generated more interest in the species and work is being done to sequence and characterize the genome of C. krusei [8].
4. Cell structure
C. krusei is usually found in one of two basic morphologies, single cell yeast or pseudohyphae; however, both forms are usually present at the same time in any growing culture [1]. In contrast to most members of the genus Candida, the cells of C. krusei are not shaped like ovals, but rather are elongated and closely resemble long grain rice when in the single-cell morphology [1]. This cell morphology is shared only with C. kejyr in the Candida genus [1]. On the other hand, when in the pseudohyphal morphology, the cells of C. krusei are elongated and attached to neighboring cells [1]. Both cell morphologies are pictured in Figure 1. The cells of C. krusei contain a cell wall that is made up of six layers that contain high concentrations of the polysaccharide alpha-D-mannan with a backbone made up of (1-6) linkages and side chains consisting of (1-2) or (1-3) linkages. These polysaccharides act as antigens on the cell surface that differentiate it from other Candida species that have different types of mannan in their cell walls [1]. Also, the C. krusei cell membrane is strongly hydrophobic which allows stronger attachment to non-polar surfaces compared to other members in the Candida genus [5]. Finally, as with most other eukaryotic cells, C. krusei cells contain organelles including ribosomes and small vesicles [1].
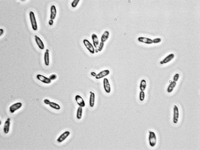
5. Metabolic processes
C. krusei is a mesophile that can only grow at temperatures up to 45 degrees C, and it is one of two Candida species that can grow in media that does not contain vitamins [1]. Like many other yeasts, it is a fermentor [9]. Under aerobic and semi-aerobic conditions, C. krusei is one of the most effective producers of ethanol from glucose compared to other members of the genus, Candida, and Pichia, a closely related genus. [9] The high ethanol yielding fermentation by C. krusei is useful in current research because buildup of organic molecules during fermentation can cause problems experimentally. For instance, C. krusei prevents the build up of succinic acid via the activity of highly expressed succinate dehydrogenase present in the Krebs Cycle. [9] Buildup of organic molecules like succinic acid can disrupt separation of ethanol from other products during ethanol production because of interference with separation methods such as pervaporation. By using this strain for ethanol fermentation, its metabolic activity makes it more suitable than other similar strains. [9]
Mechanisms of Antifungal Resistance
Anti-fungal resistance in C. krusei has been attributed to common resistance pathways in Candida species though C. krusei also presents an intrinsic resistance to fluconazole (a pyrimidine-analogue therapeutic) presented as a reduced inhibitory effect on the target enzyme Erg11p which demethylates lanosterol [10]. This loss of affinity of fluconazole for Erg11p does not seem to be caused by mutations or polymorphisms as in other Candida species [10].
C. krusei has a qualitatively different azole resistance mechanism. While most of the Candida species use a combination of ABC (ATP powered) and MFS (Proton gradient powered) multidrug transporters [11], this is not the case in C. krusei . It has a distinctive lack of MFS transporters but a broad-spectrum set of ABC transporters that contribute summatively to azole fluconazole resistance by promoting drug efflux [12]. Mechanisms of antifungals are shown in Figure 2 for reference.

Role in Chocolate Production
C. krusei is the dominant yeast present in chocolate production and cocoa bean fermentation in West African cocoa beans. Heap fermentation in particular is most effective with C. krusei [13]. Cocoa bean fermentation relies heavily on the production of ethanol from sugar in the pulp of the bean which leads to an increase in temperature during fermentation [13]. The temperature increase significantly increases the yields from cocoa bean fermentation [13]. Some strains of C. krusei were found to produce high levels of ethanol under aerobic conditions without succinic acid as a byproduct [9] likely making C. krusei an ideal yeast for heap fermentation.
6. Ecology
C. krusei is found in many places around the world [14]. Strains of C. krusei have been isolated in North America, Europe, Latin America, and the Asia-Pacific region from patient samples from over 142 hospitals [14]. In Europe, the prevalence of C. krusei is the highest among geographic regions around the world with isolates of C. krusei most common in the Czech Republic, Poland and Slovakia. In contrast, C. krusei is more uncommon in the Asia-Pacific and Latin America regions of the world [14]. Additionally, C. krusei can be isolated from a variety of food sources such as Japanese sake, Ginger Beer, and Baker’s Yeast [9]. However, C. krusei is most commonly found in patients with hematologic malignancies and recipients of blood and bone marrow transplants [14]. C. krusei susceptibility to voriconazole, a strong anti-fungal used to treat C. krusei infections, varies based on the region the C. krusei strain is localized [14]. Overall, North American strains of C. krusei had the highest rates of susceptibility to voriconazole, over 90% susceptibility. In contrast, Latin American countries had the lowest rates of susceptibility to voriconazole, with under 70% susceptibility in some countries [14].There are no clear mechanism behind these differences known but there is speculation that it could be the geographic differences in the ecology of C. krusei or the in the use of cytotoxic drugs and antimicrobial agents. [14]
7. Pathology
C. krusei is considered a multidrug-resistant pathogen due to a complex susceptibility profile and a rapid adaptive response to antifungal treatments, regardless of the drugs’ target [5]. There is a rising concern due to the spread of its infections, mostly spanning immunocompromised patients [5]. C. krusei induces Candidiasis, an infection caused by a species of the Candida genus [5]. Unlike other Candidiasis-inducing species (namely C. albicans), C. krusei has an up to 4-fold increased affinity for acrylic surfaces than epithelial cell surfaces [5]. It is hypothesized that this is the result of a strong hydrophobicity in the C. krusei cell membrane which allows stronger attachment to non-polar surfaces; these include catheter surfaces, compromising sterility and differential diagnosis. The prevalence of C. krusei candidiasis over C. albicans-induced is tied to a prophylactic oral rinse therapy that selects out C. krusei . Furthermore, C. krusei is found to be resistant to fluconazole treatment leading to negative prognosis if not noticed early on [1]. Despite the increased adherence, C. krusei remains less virulent than C. albicans unless selected for artificially [11]. Furthermore, C. krusei is more susceptible to immunological factors like lactoferrin, lysozyme, or polymorphonuclear leukocytes more so than any other Candida species analyzed, but are more resistant to murine bronchial lavage fluid [11]. This susceptibility profile indicates that immunocompromised hosts show a strong selective pressure by removing strong inhibitors while antifungals select against the common Candida species. C. krusei shares Candida-related virulence factors, such as phospholipase, proteinase and host immunological factor modulator production, phenotype switching and dimorphic transition [2].
8. Current Research
C. krusei lacks a particular focus in current research. However, the genus Candida has been seeing a renewed spotlight since Candida species are the most common human fungal pathogens [1] and infections not caused by C. albicans are on the rise [7]. Especially of note is the increased concern as of late for South American strains of C. krusei that present low susceptibility to most antifungals [6][14].
Most recently there has been research into determining why C. krusei is resistant to the antifungal fluconazole compared to similar strains such as C. albicans. Fluconazole works by inducing the formation of reactive oxidative species (ROS), however this anti-fungal induces low amounts of ROS in C. krusei cells compared to the amounts of ROS observed in C. albicans cells exposed to the drug [15]. C. krusei can activate an alternative respiratory pathway (ARP) when exposed to stress-inducing situations such as presence of an antifungal [15]. The ARP reduces the presence of ROS and decreases the rate of ROS buildup in the cell allowing time for antifungal resistant cells to develop [15]. Research towards the mechanisms of C. krusei resistance to current drugs will help develop new treatments to this emerging pathogenic bacteria.
Furthermore, the Fungal Genome Initiative along with researchers at the Broad Institute recently proposed and performed the sequencing of 5 medically relevant Candida species: C. albicans, C. tropicalis, L. elongisporus, C. guilliermondii, and C. lusitaniae, due to the fact that Candida species other than C. albicans are becoming increasingly relevant in the medical field, especially in nosocomial infections [7]. Currently, the genome of C. krusei has not been sequenced, but due to its increasing medical relevance, researchers at the University of Minnesota are beginning to attempt to sequence the organism’s genome [8].
9. References
[1] Samaranayake, Yuthika H. and Samaranayake, L.P. 1994. Candida krusei: Biology, epidemiology, pathogenicity and clinical manifestations of an emerging pathogen. J. Med. Microbiol. 41(1994), 295-310.
[2] Samaranayake, Y.H., Wu, P. C., Samaranayake, L.P., Ho, P. L. 1998. The relative pathogenicity of Candida krusei and Candida albicans in the rat oral mucosa. Journal of Medical Microbiology 47: 1047-1057.
[3] Hendriks L., Goris A., Van De Peer Y., Neefs J., VanCanneyt M., Kersters K., Hennebert G. L., and De Wachter R. 1991. Phylogenetic analysis of five medically important Candida species as deduced on the basis of small ribosomal subunit RNA sequences. Journal of General Microbiology 137: 1223-1230.
[4] Abbas, J., Bodey, G. P., Hanna, H. A., Mardani, M., Girgawy, E., Abi-Said, D., Whimbey, E., Hachem, R., Raad, I. 2000. Candida krusei Fungemia: An Escalating Serious Infection in Immunocompromised Patients. Archives of internal medicine 160(17): 2659-64.
[5] Samaranayake Y.H., Wu P. C., Samaranayake L.P., So M., Yuen K. Y. 1994. Adhesion and colonisation of Candida krusei on host surfaces. J. Med. Microbiol 41 (4): 250-258.
[6] Ricardo E. T. A. 2015. Genetic and Molecular Insights of Candida Krusei Antifungal Resistance. Porto, Faculdade De Medicina Da Universidade Do Porto.
[7] Butler, Geraldine et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida
genomes. Nature 459: 657-662
[8] Genomic Analysis of Candida krusei. 2016. University of Minnesota: Minnesota Supercomputing Institute;
[2016; November 29, 2016]. https://www.msi.umn.edu/content/genomic-analysis-candida-krusei
[9] Nakayama S., Morita T., Negishi H., Ikegami T., Sakaki K., and Kitamoto D. 2008. Candida krusei produces ethanol without production of succinic acid; a potential advantage for ethanol recovery by pervaporation membrane separation. FEMS Yeast Research 8(5). 706-714.
[10] Morio, F., Loge, C., Besse, B., Hennequin, C., Le Pape, P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical Isolates: new substitutions and a review of the literature. Diagnosis of Microbiological Infectious Diseases 66: 373-384.
[11] Panwar, S.L., Pasrija, R., Prasad, R. 2008. Membrane homeostasis and multidrug resistance in yeast. Bioscience Reports 28: 217.
[12] Gaur, M., Puri, N., Manoharlal, R., Rai, V., Mukhopadhayay, G., Choudhury, D., Prasad, R. 2008. MFS transportome of the human pathogenic yeast Candida albicans. BMC Genomics 9: 579.
[13] Jesperson L., Nielson D.S., Hønholt S., Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Research 5 (4-5): 441-453.
[14] Pfaller, M.A., Diekema, D. J., Gibbs, D. L., Newell, V. A., Nagy, E., Dobiasova, S., Rinaldi, M., Barton, R., Veselov, A., and the Global Antifungal Surveillance Group. 2008. Candida krusei, a Multidrug-Resistant Opportunistic Fungal Pathogen: Geographic and Temporal Trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. Journal of Clinical Microbiology 46(2): 515–521.
[15] Costa‐de‐Oliveira S., Sampaio‐Marques B., Barbosa M., Ricardo E., Pina‐vaz C., Ludovico P., Rodrigues A. 2012. An Alternative Respiratory Pathway on Candida krusei: Implications on Susceptibility Profile and
Oxidative Stress. FEMS Yeast Research 12 (4):423-429
[16] Candida Krusei. 2014. UC Davis:
Viticulture and Enology; [2016; November 29, 2016]. http://wineserver.ucdavis.edu/industry/enology/winemicro/wineyeast/candida_krusei.html
Edited by Kailyn Doiron, Beth Grinkevich, Alvaro Dafonte Imedio, and Puja Patel students of Jennifer Talbot for BI 311 General Microbiology, 2016, Boston University.
