Chlorobium tepidum
A Microbial Biorealm page on the genus Chlorobium tepidum
Classification
Higher order taxa
Domain: Bacteria; Phylum: Chlorobi; Class: Chlorobia; Order: Chlorobiales; Family: Chlorobiaceae
Species
Genus: Chlorobaculum; Species: Chlorobium tepidum
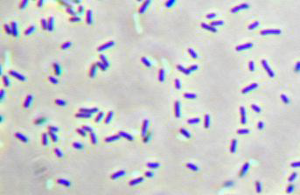
Description and significance
Chlorobium tepidum is an anaerobic phototrophic bacterium that is also a thermophile. As a member of the Chlorobiaceae family, it is a model organism of green sulfur bacteria. (4) The habitats of this bacterium include anoxic and sulfide-rich waters, mud, sediments, microbial mats, and even microbial consortia. (4) The cells of this bacterium are gram-negative nonmotile rods of varying lengths that photooxidize reduced sulfur compounds. (5) C. tepidum was isolated from certain acidic high sulfide New Zealand hot springs. (5) Under photoautotrophic conditions, the growth rate of this bacterium is faster than any other anaerobic phototroph as generations are produced about every two hours. (1) Consequently, it is an ideal candidate for studying photosynthesis and autotrophy in green bacteria.
Genome structure
The genome of C. tepidum has been completely sequenced. Its genome is one circular chromosome that consists of 2,154,946 basepairs. (1) Through genome comparisons, it has been identified that many genes in C. tepidum are highly conserved among photosynthetic species, and though they have no clear function, they may play important roles in photosynthesis or photobiology. (1) In addition, phylogenomic analysis has revealed probable duplications of genes that are involved in biosynthetic pathways for photosynthesis and the metabolism of sulfur and nitrogen. (1) Consequently, strong similarities have been discovered between the metabolic processes of C. tepidum and many Archaeal species. (1)
Cell structure and metabolism
C. tepidum is a gram-negative cell that harvests light through special photosynthetic systems known as chlorosomes. (4) Chlorosomes contain highly aggregated light-harvesting centers, including bacteriochlorophyll c and carotenoids that are surrounded by a protein-stabilized galactolipid monolayer that lies at the inner surface of the cytoplasmic membrane. (4) Two unusual features of this bacterium are its inability to produce oxygen and its low toleration of oxygen despite being a photosynthetic organism. In addition, as a member of the phylum Chlorobi, C. tepidum has the ability to fix carbon dioxide using a reverse tricarboxylic acid cycle using electrons from hydrogen or reduced sulfur. (1)
Ecology
Due to their anaerobically photosynthetic nature, C. tepidum and other green sulfur bacteria have been suggested to be among the first photosynthetic organisms that arose during a time when there was a low amount of oxygen present on Earth. Consequently, C. tepidum can be found in anoxic and sulfide-rich waters, mud, sediments, and microbial mats. (4) The optimum temperature for such bacterium to grow ranges from 47–48°C. C. tepidum can transform sulfur available in the environment around it into a form that can be absorbed by other living organisms. In other words, C. tepidum is directly involved in the global sulfur cycle. In addition, since C. tepidum can metabolize nitrogen, it is also a player in the world's nitrogen cycle.
Pathology
C. tepidum is currently not known to cause diseases.
Application to Biotechnology
As the first from the phylum Chlorobi to have its genome completely sequenced, C. tepidum is a model organism for scientists studying green sulfur bacteria. (4) Through the use of its genome sequence, scientists are studying the origin and mechanisms of photosynthesis. Consequently, because C. tepidum harvests light for energy, scientists are hoping that the bacterium could one day serve as an alternative source of energy. (6) In the process of photosynthesis, C. tepidum metabolizes nitrogen and sulfur, two critical nutrients for cells. The bacterium's usage of nitrogen and sulfur compounds translates to its involvement in the global cycles of both nutrients, and as a result, scientists are hoping to understand the diverse ways that these elements are processed in individual cells. By doing so, scientists believe they will be able to discover information that will allow them to produce biofuels in the future. (6)
Current Research
Marine biologist Thomas Hanson investigates how microorganisms are able to survive and compete in the environment. (2) Instead of focusing on the negative aspects of bacteria, Hanson has chosen to study how microorganisms contribute to the maintenance of favorable conditions for life. (2) Hanson has decided to identify and characterize the genes and proteins present in C. tepidum and how they contribute to the bacterium’s ability to transform sulfur compounds into usable forms for other organisms. (2) By determining the bacterium's sulfur oxidation reactions, Hanson's project will give more information about the relatively poorly understood global sulfur cycle.
Donald A. Bryant's laboratory focuses on the chlorosome, the light-harvesting organelle found in the C. tepidum that consists mainly of bacteriochlorophyll c. Through the process of homologous recombination, specific genes in the bacterium can be targeted for inactivation. (4) Consequently, more than 30 mutants have been developed by this process, allowing Bryant and his lab to gain more insight about the proteins found in the chlorosomes as well as the biosynthetic pathways of bacteriochlorophylls and carotenoids. (4)
Thomas E. Hanson and F. Robert Tabita have discovered a gene encoding a product that is similar to ribulose-1,5-bisphosphate carboxylase/oxygenase, also known as RubisCO, in the genome sequence of the C. tepidum. (3) In order to gain information about the RubisCO-like proteins, the C. tepidum gene encoding the RubisCO-like protein was disrupted. Consequently, the resulting mutant strain had defects in photopigment content, photoautotrophic growth, carbon fixation rates, and sulfur metabolism. (3) In addition, the mutant strain showed an accumulation of two oxidative stress proteins. (3) With these results, C. tepidum's RubisCO-like protein is thought to be involved in oxidative stress responses and sulfur metabolism, giving way to the belief that the protein could be an evolutionally link to RubisCO. (3)
References
2. Gulbrandsen, Kari. "Marine biologist wins top NSF grant." Delaware EPSCoR
4. PennState: Research on Chlorobium tepidum in the Bryant laboratory
6. Winstead, Edward R. "Light-harvesting bacterium C. tepidum is sequenced." Genome News Network
Edited by student of Rachel Larsen and Kit Pogliano
