Circovirus
A Viral Biorealm page on the genus Circovirus
Baltimore Classification
Higher order taxa
Viruses; ssDNA viruses; Circoviridae; Circovirus
Species
Beak and feather disease virus, Porcine circovirus 1, Porcine Circovirus 2, Canary circovirus, Goose circovirus, seven unclassified circoviruses
Description and Significance
Circoviruses are very small non-enveloped icosahedral viruses with a single stranded circular DNA genome. They have been described in pigs (porcine circovirus, PCV), chickens (chicken anemia virua, ChAV), psittacines (psittacine beak and feather disease virus, PBFD) and pigeons. The virus targets the lymphoid tissue and causes immunosuppression in the host.
Circoviruses are the smallest pathogenic DNA viruses that have been identified and characterized in animals.
Genome Structure
The genome of the circovirus is monomeric. It is unsegmented and contains a single molecule of circular, ambisense, single-stranded DNA that forms a covalently closed circle. The complete genome is remarkably small, often containing just two major genes: rep (encoding the replicase protein) and cap (encoding the capsid protein). The rep genes may however generate variant products. The genome is typically 1800-2000 nucleotides long and the genome has a guanine+cytosine content of 48.4- 53.1 %. (source: ICTV dB Descriptions) For an example of genetic study of circoviruses, see Hughes and Piontkivska, 2009.
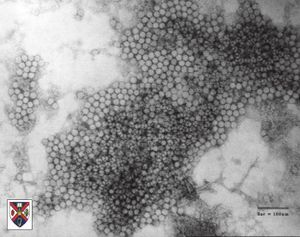
Virion Structure of a Circovirus
The virions of a circovirus consist of a capsid with no envelope. Virions have an extracellular phase during their life cycle. The virions are not tailed. The capsid is round and exhibits icosahedral symmetry (T=13). It is isometric and has a diameter of 17-22 nm, although the size of particle diameter varies with the type of stain used. The capsids appear round and the capsid surface structure reveals a regular pattern with distinctive features. The capsid consists of 32 capsomers. The capsomers are hollow morphological subunits. (source: ICTV dB Descriptions)
Reproduction Cycle of a Circovirus in a Host Cell
In case of the Porcine circovirus type 1 (PCV1), the rep gene directs the synthesis of two proteins. The full length protein Rep is 312 amino acids in size, the spliced variant Rep' is truncated and exon 2 is frame-shifted. The replication on PCV1 DNA depends on the synthesis of both proteins. Rep and Rep' bind in vitro to double-stranded DNA fragments comprising part of the origin of replication of PCV1, but the minimal binding sites of the two proteins are distinct. Rep protein represses the promoter of the rep gene by binding to the two inner hexamers H1 and H2. Although Rep' bind to the same sequence, it does not influence Prep. The ratio of the two transcripts varies later on, although similar amounts of rep and rep' are detected by real time PCR immediately after PCV1 infection. Both proteins are co-localised in the nucleus and the formation of homo- and heteromeric complexes is also possible.
Viral Ecology & Pathology
References
Austin L. Hughes and Helen Piontkivska. 2008. Nucleotide Sequence Polymorphism in Circoviruses. Infect. Genet. Evol. 8:130–138.
Leslie W. Woods, DVM, PhD, Dipl ACVP, and Kenneth S. Latimer, DVM, PhD, Dipl ACVP; "Circovirus Infection of Nonpsittacine Birds". 2000. Journal of Avian Medicine and Surgery 14(3):154–163.
Annette Mankertz, Rifat Çaliskan, Kim Hattermann, Bernd Hillenbrand, Petra Kurzendoerfer, Bettina Mueller, Cornelia Schmitt, Tobias Steinfeldt, Tim Finsterbusch. 2004. "Molecular biology of Porcine circovirus: analyses of gene expression and viral replication". Veterinary Microbiology 98:81–88.
