Clostridium magnum
Classification
Classification 7
Domain: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Clostridiales
Family: Clostridiaceae
Species: Clostridium magnum
Description and Significance
The name "Clostridium magnum" comes from the Greek "kloster" meaning "spindle" 4 and the latin "magnus" meaning "great" 5 which is explained by observing the size of the bacteria since its dimensions are 1.0-4.0um x 4-16um. 3 Clostridium magnum is rod shaped and slightly pointed at the ends. 3 C. magnum is gram negative 3 however it may also appear as gram positive 2 due to it's cell structure being much more similar to gram positive with a thin murein layer.3 C. magnum is also motile by polar and sub polar flagella, strictly anaerobic, and acetogenic.3 The habitat of Clostridium magnum is in anoxic freshwater sediments and digestor sludge.3
Clostridium magnum is an acetogen which indicates it plays an important role in binding Hydrogen molecules which would otherwise seize up the process of fermentation. Through acetogenesis, C. magnum is able to take excess H2 molecules and produce acetate.
Genome Structure
The genome of C. magnum has not been completely determined and there is no current project to determine its genome.6 However, some genes have been sequenced as well as a partial sequence of the 16S rRNA.2 Some genes that were sequenced are the acoR, acoA, acoB, acoC, acoL, and the acoX gene; specifically these genes are a part of the acetoin dehydrogenase enzyme system.8
Cell Structure, Metabolism and Life Cycle
An outer membrane layer was not observed when using an electron microscope in ultrathin sections as is typical in Gram-negative bacteria.3
C. magnum is a strict anaerobe chemoorganotroph that can use many different sugars and acids: "[f]ructose, glucose, sucroe, xylose, malate, citrate, 2,3-butanediol, acetoin, and (weakly) pyruvate".3 It cannot grow on many C1 compounds that other homoacetogenic Clostridium can grow on, such as: "ethylene glycol, ethanol, acetate, glyoxylate, glycolate, serine, lactate, oxalate, malonate, fumarate, succinate, oxaloacetate, glutamate, glycerate, glycerol, diacetyl, orotate, maltose, ribose, arabinose, starch, peptone, casamino acids, yeast extract". 3 In addition, "[g]elatin or urea [wasn't] hydrolyzed" and "[n]itrate, sulfate, sulfite, sulfur, thiosulfate or fumarate [was] not reduced" (254).3 After observing C. magnum, the only product found from fermentation was acetate regardless of the substrate provided (unless it was C1 compounds, then no growth was found).3 Schink determined the enzymes that participated in butanediol degradation included "[a]cetoin dehydrogenase, phosphate acetyl transferase and acetate kinase, whereas no butanediol dehydrogenase activity could be detected".3 It is thought that butanediol is broken down "via cleavage into two C2 units" and then is "oxidized to acetate with concomitant ATP synthesis" (252). 3
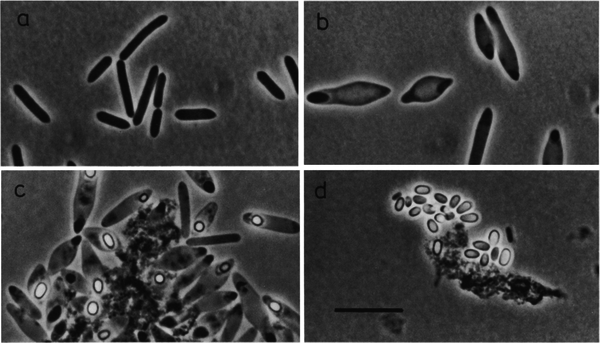
When growing C. magnum on "glucose or fructose, cells swell in the middle and became spindle-like" (250).3 Also, "[d]ark elliptical zones could be observed at the spindle tips" and "[s]pore formation occurred in the central to subterminal region" as shown in Figure 1c. 3 The spores that were formed "were elliptical…and were released only very late after growth on glucose", shown in Figure 1d.3 In order for the spores to germinate "in fresh media", the "addition of a trace [amount] of sodium dithionite" was required (251).3
Ecology and Pathogenesis
Due to C. magnum's abilities to convert carbon gases to acetic acid, there have been proposals to release C. magnum into the environment in New Zealand to test for it's environmental impact and potential implications for acetic acid in industrial applications. In the proposal, the identification and assessment of C. magnum as a pathogen to humans, plants, and/or animals was assessed along with the likelihood of C. magnum's impact on the genetic diversity of the indigenous species of New Zealand. 1 Although under certain conditions C. magnum is capable of forming endospores, "[a]fter reviewing the known biological characteristics, there is no evidence that C. magnum will cause disease in humans, plants and animals or will become a vector for human, plant or animal diseases." 1 C. magnum is likely to possess biogeographical characteristics in the sense that it is a strict anaerobe but as a member of the Clostridium genus, it is also likely to act as a saprobe (meaning they live on decaying organic matter); in the case that C. magnum acts as a saprobe, it is probable that it will grow ubiquitously.1 Supposing C. magnum does grow ubiquitously it may not actually be metabolically active unless the environment C. magnum finds itself in has appropriate conditions for metabolic processes; these conditions include but are not limited to, "temperature, soil texture and structure, pH, salinity, moisture levels, which plants are grown in the soil, and the presence or absence of suitable resources." 1 In conclusion, "there is no evidence that C. magnum will have adverse effects on the environment or human health and safety, and whether C. magnum can or cannot produce spores does not change this assessment." 1
References
5) "magnum." Dictionary.com Unabridged. Random House, Inc. 09 Apr. 2014.
Author
Page authored by Samantha Hoyle and Chelsea Ball, students of Prof. Ned Walker and Prof. Kaz Kashefi at Michigan State University.
