Conexibacter
Classification
Domain: Bacteria
Phylum: Actinobaceria
Class: Actinobaceria
Sub-Class: Rubrobacteridae
Order: Solirubrobacteriales
Family: Conexibacteraceae
Genus: Conexibacter
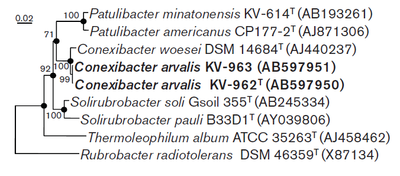
Species
Conexibacter woesei (DSM 14684) (type strain), Conexibacter arvalis (DSM 23288)
Description
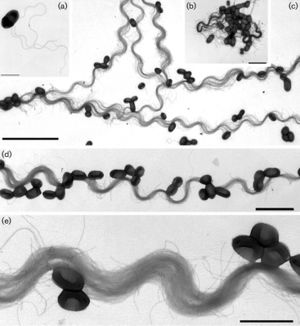
Bacteria of the genus Conexibacter are characterized as small rods with width of 0.6-0.7 µm and length of 0.9-1.2 µm. They are Gram-positive, aerobic, non-sporulating and are motile by characteristic long peritrichous flagella. The type strain of this genus is Conexibacter woesei named in honor of Carl R. Woese, a pioneer with his work on the use of 16S rRNA for phylogenetic analysis. C. woesei was isolated from forest soil in Gerenzano, Italy and grows optimally at pH 7-7.5 and at 28-37ºC. [1]
The genus Conexibacter was first developed in 2003 with the discovery of the type strain C. woesei.[1] In 2012 a second species, Conexibacter arvalis, was isolated from Japanese field soil. Both isolates are available in pure culture. The genus is known as a very deep branch of the phylum Actinobacteria having only a 90% similarity to the genus Thermoleiphilum, and being only 83.8% similar to the well known genus Rubrobacter, which contains species known to be the only thermophiles that are radiation resistant.[2][5][6] Since it is such a deep reaching branch of Actinobacteria there is not much diversity in the lineage of Conexibacter. This lack of diversity is further amplified by the presence of only two known species. However C. wosesi and C. arvalis do represent some diversity between them. Specifically C. woesei was found to be oxidase positive while C. arvalis is oxidase negative, suggesting that C. arvalis may have an alternative electron acceptor to C.woesei or that it may have use a different cytochrome to transfer terminal electrons to oxygen. C. arvalis has a higher G+C content than C. woesi with 74-5% G+C compared to 71% G+C and it is slightly more halotolerant.[1][5]
The genus Conexibacter is a soil bacteria that is not primarily studied. Other than the discovery of two isolates, these species are only reported when they are discovered in metagenomic studies. However, though not much is known about this genus some characteristics of it's species make them very intriguing. Once such example is their chemotaxonomy. Both C. wosesi and C. arvalis have the tetrahydrogenated menaquinone MK-7(H4) as the major component of their plasma membrane, a trait that has not yet been reported in bacteria. Another intriguing feature of these species is their long, peritrichous flagella. It was found that if cultures of C. woesei were allowed to grow for over a week the long flagella tend to become entangled forming long, rigid, non-motile spiral structures.[1][5]
Ecology and Significance
Though Conexibacter species have not been extensively studied, there are a few studies that have provided hints to its significance in both its environment and for human utilization. Conexibacter are strictly aerobic and can reduce nitrate to nitrite which suggests they may play a role in the nitrification process, especially in oxygen limiting states. Their ability to reduce nitrate is also what makes them most intriguing for industrial applications such as bio-remediation. However due to their slow growth rate there are likely better suited organisms available. In addition potentially aiding in the nitrogen cycling of soil they are also characterized as saccharolytic, meaning they get their carbon and energy primarily from carbohydrate hydrolysis. This ability indicates that they likely contribute to carbon cycling in soil ecosystems.[1][2][5]
Conexibacter have been identified by isolation from Italian forest soil and Japanese field soil.[1][5] In addition to isolation, there have been a number of metagenomic studies that have identified the presence of Conexibater species in many various environments. These environments include soil, soil crusts, sediment, rhizosphere, acidic Sphagnum peat bog, and permafrost from Russia and Alaska.[2][4] Though their ideal temperature ranges were reported to be 28-37ºC both C. woesei and C. arvalis were reported to grow at temperatures as low was 4ºC. In the Alaskan permafrost metagenomic study it was found that a Conexibacter was, with Spartobacteria, the most dominant genera in a long-thawed sample of tundra located in the frozen portion of the soil organic horizon. Their role in this environment is unknown however it is believed that their slow growth and high G+C content may factor into their survival at such harsh conditions. It is predicted that they are contributing to carbon cycling in soil. Interestingly, the layer of the soil that Conexibacter was found to be dominant was also the most nitrogen rich as well, suggesting that its ability to reduce soil nitrates may provide a competitive edge in certain environmental conditions.[3]
The genus Conexibacter is a very intriguing genus with organisms that have unique capabilities. They are believed to contribute to carbon cycling in soil environments and have the potential to also be contributing to nitrogen cycling. The nitrate reduction ability combined with their general hardiness makes them unique candidates from nitrate bio-remediation in psychotropic environments once more is understood about their metabolic capabilities. Though they are believed to have little significance at present, with more research they may play a significant role in bio-remediation in the future.
Genome Structure
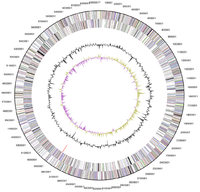
The complete genome of Conexibacter woesei was published in 2010. The genome size is 6,359,369 bp with a DNA G+C content of 72.73%. There are 5,998 genes with 5,950 of them being protein coding genes. A few protein coding genes to note are multiple nitrate reductases, nitrate transporters, and multiple antibiotic and antibiotic-resistance genes in addition to general carbohydrate degradation pathway genes. This supports the literature claim that this isolate is capable of nitrate reduction. Another interesting thing about this genome is the presence of genes coding for cytochrome d ubiquinol oxidase, in addition to the standard aerobic cytochrome c oxidase coding genes. The cytochrome d ubiquinol oxidase coding genes may be conserved in C. arvalis, providing a potential explanation to its negative oxidase test result. The genome additionally describes potential gene products having the capacity of drug resistance, symbiosis, and opportunistic pathogenicity.[1][2][5]
Metabolism
Conexibacter species are characterized as strictly aerobic chemoorganotrophs. They are saccharolytic, utilizing the hydrolysis of carbohydrates as their primary source of energy and carbon.[1] They are known to be very slow growing, with C. arvalis taking 14 days to sufficiently cultivate in pure culture.[5] This may be indicative of their type of metabolism and could suggest that they may be dependent on symbiotic organisms for the degradation of more complex materials to increase carbohydrate availability. One interesting feature present in both species is the ability to reduce nitrate into nitrite.[1][5] This ability, coupled with the lack of cytochrome c oxidase in C. arvalis may suggest the possibility of anaerobic respiration occurring in oxygen limiting conditions using nitrate as the terminal electron acceptor. More research on the genus must be preformed to verify the activity of potential pathways described in the C. woesei genome.
References
[1] Monciardini Palo, et al. 2003. Conexibacter woesei gen. nov., sp. nov. a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int J Syst Bacteriol. 53:569-576.
[2] Pukall Rüdiger, et al. 2010. Complete genome sequence ofConexibacter woeseitype strain (ID131577). Standards in Genomic Studies. 2:212-219.
[3] Deng Jie, et al. 2015. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in Alaska. Molecular Ecology. 24:222-234.
[4] Krivushin Kirill, et al. 2015. Two Metagenomes from Late Pleistocene Northeast Siberian Permafrost. Genome Announc. 3(1):01380-14.
[5] Seki Tamae, et al. 2012. Conexibacter arvalis sp. nov., isolated from a cultivated field soil sample. Int J Syst Bacteriol. 24:222-234.
[6] Ferreira Anna, et al. 1999. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles. 3:235-238.
Author
Page authored by Andrew Steinberger, student of Prof. Katherine Mcmahon at University of Wisconsin - Madison.
