Deinococcus radiodurans: Extreme Radiation Tolerance
Classification
Higher Order Taxa
Bacteria (Domain); Deinococcus-Thermus (Phylum); Deinococcus (Class); Deinococcales (Order); Deinococcaceae (Family); Deinococcus (Genus)
Species
radiodurans
Description
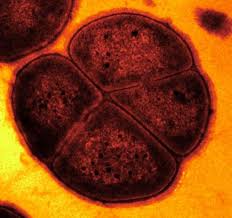
Deinococcus radiodurans was originally discovered in 1956 by Anderson et al. who noticed the presence of bacteria in a can of ground beef that was sterilized by gamma irradiation [8]. Deinococcus radiodurans was first classified as Micrococcus radiodurans, due to its similarities to that genus. However, after further research, it was later put into a phylum of its own [4]. “Deinos”, in Greek, means strange or unusual, which is a good description of this bacteria [7]. It is a spherical, non-motile, non-spore forming bacterium with a red pigment [6]. This gram positive bacteria is 1.5-3.5 μm in diameter and often forms tetrads but can also be found in singles of pairs [6]. Its cellular envelope more closely resembles that of a gram negative bacterium as it has 6 layers- (from inside out) cell membrane, peptidoglycan, compartmentalized layer, electron transport zone, outer membrane and a hexagonally packed intermediate layer [3]. The genome of D. radiodurans was sequenced and published in 1999 by TIGR. D. radiodurans is novel due to its ability to survive in extremely high levels of radiation [8].
Ecology and Significance
The natural environment of Deinococcus radiodurans is not well defined [1]. It has been isolated around the world in places with high concentrations of organic nutrients, such as in animal feces, soil, sewage, and processed meats [6]. They also have been isolated from Fukushima Daiichi and Chernobyl both locations of nuclear accidents [2]. There are four species that make up the Deinococcus genus: Deinococcus radiodurans, Deinococcus proteolyticus, Deinococcus radiopugnans, and Deinococcus radiophilus [6]. The Deinoccoceae lineage all have a degree of radiation resistance but also may individually be a mesophile, thermophile, psychrophile, or a combination of more than one, surviving in environments from hot springs, to deserts, to Antarctica [7]. Out of these, D. radiodurans is the most extreme in terms of radiation resistance and desiccation tolerance [1]. Cyanobacteria such as Chroococcidiopsis and fungi such as Filobasidiales also have the characteristic of radiation resistance but not to the extent of D. radiodurans [7]. D10, the dose required to kill 90% of population is 0.005 kGy for humans, 0.25 kGy for Escherichia coli and can be as high as around 10 kGy for D. radiodurans [1]. There are no natural environments that have radiation greater than 400 mGy per year so their resistance is more likely due to the fact that they have genes that allow them to survive desiccation, which causes double-stranded breaks [4].
Genome Structure and Mechanisms of Resistance
Radiation is caused by electromagnetic waves and the results on a cell are oxidation damage to proteins, DNA, RNA, and enzymes [2]. One of the main causes of cell death is due to the double stranded breaks (DSBs) that occur in the DNA [4]. Unlike most other bacteria, D. radiodurans retains its viability in radiation. It can withstand up to 21,000 J/m2 of ultraviolet radiation which usually results in cyclobutane dimers and pyrimidine-pyrimidone photoproducts which cause CC-TT or C-T base pairing Schematic of origin of different types of radiation and its effects on extremophiles. These bacteria can also withstand 5 kGy of Gamma radiation without loss of viability [4.
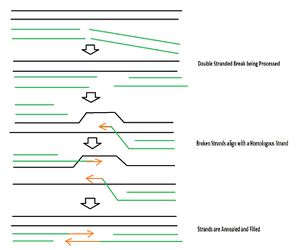
D. radiodurans does not have a mechanism to prevent DSBs, it only has mechanisms to repair the DNA after these breaks have occurred [4]. The genome is in a condensed ring to prevent diffusion of the DSBs [4]. Following exposure to radiation, cellular regulatory mechanisms prevent replication of DNA [4]. Proteins called ddrA and pprA then bind to the ends of all the DNA breaks to protect them from degradation by nucleases [5]. The DSB ends are then processed by UvrD and Rec J into 3’ single strands of DNA [7]. RecFOR loads RecA onto DSBs which searches for double stranded DNA homology and also primes DNA repair synthesis on partially overlapping fragments of the homologous DSB which are to be used as templates [7]. The 3’ ends of a DSB goes in between strands of a matching unbroken strand and each end is base paired by Pol A and then joins the other annealing strand [4] (see Figure 3).
Recombinational Repair is possible due to the fact that this bacterium has a segmented genome and has 4-10 copies of the genome in each cell [4]. Along with making it very unlikely that every copy will be severely damaged, it can use the multiple copies to homologously recombine, repairing its genome [5]. RecA is the protein used for dependant homologous recombination [10]
Mn (II) is also required for the cells survival as it facilitates the cleanup of Reactive Oxygen Species (ROS) which are produced during radiation [7]. These ROS are what damage lipids, proteins, nucleic acids, carbohydrates and create DSBs [7].
Current Research and Applications in Biotechnology
A recent application of D. radiodurans is to utilize them for bioremediation [2]. At sites of nuclear weapon production, there are many radioactive wastes such as Mercury (Hg) (II), 235-Uranium, and toluene all which are highly toxic [5]. D. radiodurans is able to aid in Mercuric Reductase bioremediation by reducing Hg (II) to less toxic elemental mercury (Hg (0)) [5]. In these places of nuclear waste, the radiation can be as much as 10 mCi/L and the cost to clean these sites can be as much as 265 billion dollars [5]. To use microbes for bioremediation, a bacteria needs to be able to break down the toxic agents and also have the ability to survive in a highly radioactive environment [5]. E. coli BL308 has a mercuric reductase gene called merA that reduces Hg (II) to Hg (0) but E. coli cannot survive in high radioactivity [5]. On the other hand, D. radiodurans is not affected by radiation but does not have a gene for mercuric reductase [5]. Scientists have been able to isolate the merA gene and clone it into D. radiodurans, creating a microbe that has both the characteristics needed for this particular bioremediation process [5]. In another study, Appukuttan (2011) cloned a gene called PhoN which allows cells to precipitate Uranium from nuclear waste sites [9].
There are also studies looking into exploiting the radio-resistant mechanisms for cancer prevention and skin protection [2].
References
(1) Das, A., D and Misra, H. “Hypothetical proteins present during recovery phase of radiation resistant bacterium Deinococcus radiodurans are under purifying selection”. Journal of molecular evolution, 2013, Volume 77, Issue 1, pp. 31 – 42.
(2) Gabani, P., Singh, O. “Radiation-resistant extremophiles and their potential in biotechnology and therapeutics”. Applied Microbiology and Biotechnology, 2013, DOI: 10.1007/s00253-012-4642-7
(3) Misra, H. “Physiological and molecular basis of extreme radioresistance in Deinococcus radiodurans”. Current science (Bangalore), 2013, ISSN: 0011-3891.
(4) Cox, M. “Deinococcus radiodurans - the consummate survivor.” Nature reviews, Microbiology, 2005, DOI: 10.1038/nrmicro1264.
(5) Brim, H. “Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments”. Nature biotechnology, 2000, DOI:10.1038/71986.
(6) Battista, J. "Against all odds: the survival strategies of Deinococcus radiodurans". Annual review of microbiology, 1997, Volume 51, p. 203.
(7) Slade, D., and Radman, M. “Oxidative stress resistance in Deinococcus radiodurans”. Microbiology and molecular biology reviews, 2011, Volume 75, Issue 1, pp. 133 - 191.
(8) Anderson, A., Nordon, H., Cain, R., Parrish, G., and Duggan, D. “Studies on a Radio-Resistant Micrococcus. I. Isolation, Morphology, Cultural Characteristics, and Resistance to Gamma Radiation”. Food Technology, 1956, Vol. 10, Issue 2, p. 575.
(9) 9. Appukuttan, D. “PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation”. Journal of biotechnology, 2011, DOI:10.1016/j.jbiotec.2011.05.002.
(10) Carroll, J., Daly, M. and Minton, K. “Expression of recA in Deinococcus radiodurans”. Journal of Bacteriology, 1996, Volume 178, Issue 1, pp. 130 - 135.