Desulfatibacillum alkenivorans
Classification
Higher order taxa
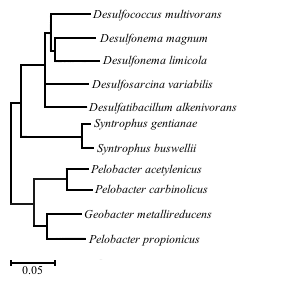
Bacteria; Proteobacteria; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfatibacillum
Species
Desulfatibacillum alkenivorans AK-01
Description and Significance
Desulfatibacillum alkenivorans AK-01 is a specific strain of D. alkenivorans isolated from estuarine sediments at the Arthur Kill waterway bordering New Jersey, a site chronically contaminated with petroleum [10]. Analysis showed that AK-01 is metabolically versatile, capable of coupling sulfate reduction with the oxidation of certain alkanes, alkenes, organic acids, and fatty acids [11]. The ability of AK-01 to grow on alkanes has significant implications for bioremediation of anoxic sites that have been heavily polluted with petroleum [3]. The recalcitrance and insolubility of hydrocarbons hinders remediation efforts [1]. Alkanes require activation by ultraviolet radiation, high temperatures or chemical catalysts [1]. In the absence of these conditions, biotransformation becomes one of the most important processes of alkane removal [2]. D. alkenivorans AK-01 serves as a model organism for the study of anaerobic alkane biodegradation with implications in bioremediation and the global carbon cycle [3]. To date, only ten other anaerobic bacteria capable of degrading alkanes have been described in literature. Bacteria coupling alkane degradation with nitrate reduction all belong to the Betaproteobacteria and Deltaproteobacteria [15]. The bacteria capable of coupling sulfate reduction with alkane degradation belong to the Deltaproteobacteria [15]. Of these, Desulfococcus oleovorans Hxd3 is the only other organism with a sequenced genome [12], further demonstrating the value of D. alkenivorans as a model organism [3].
Genome Structure
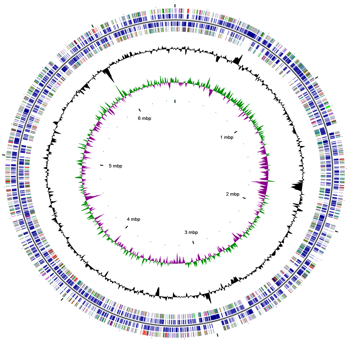
The complete genome of D. alkenivorans AK-01 has been sequenced by the U.S. Department of Energy (DOE) Joint Genome Institute. The genome consists of a circular chromosome containing 6.5 Mbp that encode 5361 genes [13]. 5296 of the genes encode proteins, although the function of only 68.51% of these proteins has been predicted [13]. More than 100 genes are directly related to flagellum structure, chemotactic mechanisms, and the assembly of the flagellum and pilus [3]. The discovery of these genes suggests that motility may be expressed under specific environmental condition when it becomes a selective advantage [3]. Thus far, however, motility in D. alkenivorans AK-01 has not been detected [10], nor has the presence of a flagellum under typical growth conditions on alkanes [11]. The presence of genes involved in the reduction of oxygen and the tolerance of toxic oxygen species also indicates that AK-01 could be adapted to the boundary between oxic and anoxic environments [3].
Cell structure, Metabolism and Life cycle
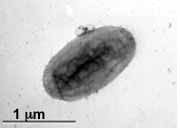
Desulfatibacillum alkenivorans AK-01 is a short, non-sporulating, gram-negative rod that is approximately 1.5 μm in length and 0.5 μm in diameter [10]. Strain AK-01 is mesophilic. It grows optimally at temperatures between 26°C and 28°C [13]. The optimal pH is between 6.9 and 7, whereas the optimal NaCl concentration is approximately 10 grams/L [10]. The doubling times of AK-01 at optimal conditions were 0.8, 1.2, and 3 days with hexadecanoate, octanoate, and hexadecane as growth substrates, respectively [10]. D. alkenivorans AK-01 is capable of three forms of metabolism including heterotrophy, autotrophy, and syntrophic alkane metabolism [3].
Autotrophy
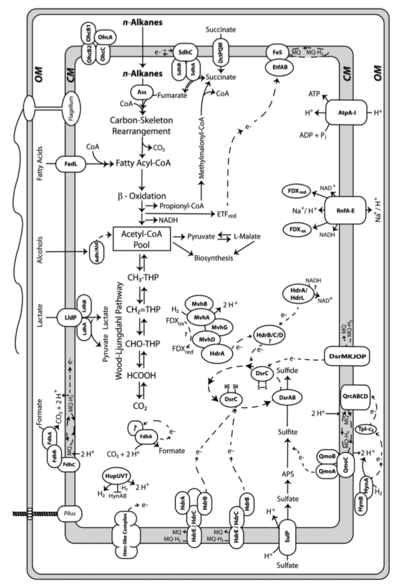
Desulfatibacillum alkenivorans AK-01 can grow chemolithoautotrophically with H2, CO2, and sulfate [10]. The Wood-Ljungdahl pathway is the predicted mechanism of CO2 fixation due the presence of all the required genes for the pathway and the absence of the gene for ATP citrate lyase, an important enzyme within the reverse TCA cycle [3]. The Wood-Ljungdahl pathway is used to synthesize cellular carbon from CO2 while obtaining energy from the coupling of hydrogen oxidation to sulfate reduction [3].
Syntrophy
Desulfatibacillum alkenivorans AK-01 is capable of syntrophically metabolizing alkanes when coupled with Methanospirillum hungatei JF-1. M. hungatei is an anaerobic methanogen that utilizes formate or H2-CO2 [7]. When AK-01 was grown with M. hungatei JF-1 in a medium free of sulfate, methane was produced from n-hexadecane [3]. Cultures of M. hungatei JF-1 and D. alkenivorans AK-01 grown separately failed to produce methane [3]. The mechanism of syntrophic metabolism was predicted after the detection of three formate dehydrogenase complexes in the AK-01 genome. The formate dehydrogenase complexes are capable of producing formate from CO2 and H2, using electrons from a menaquinone pool produced during alkane metabolism. This suggests that formate production may be utilized to transfer electrons derived from alkanes to the methanogen [3].
Heterotrophy
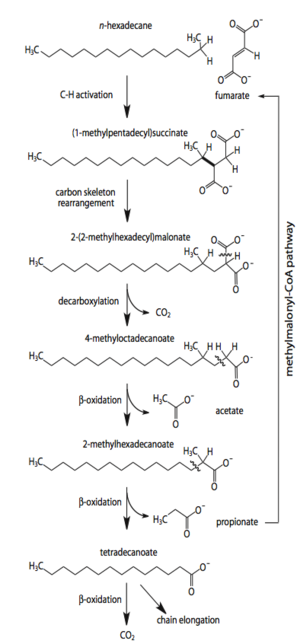
Desulfatibacillum alkenivorans AK-01 is capable of using sulfate, sulfite and thiosulfate, but not sulfur, as terminal electron acceptors during heterotrophic metabolism [10]. Alkane degradation is initiated through the subterminal addition of the alkane carbon to the double bond of fumarate [2]. The alkylsuccinate intermediate is further degraded through processes of carbon-skeleton rearrangement, decarboxylation, and beta-oxidation [1]. The resultant acetate molecule is further oxidized to CO2 through the reversal of the Wood-Ljungdahl pathway [3]. Strain AK-01 is able to completely oxidize alkanes to carbon dioxide when coupled with sulfate reduction [11]. This was shown through the formation of radiolabeled CO2 from radiolabeled [1-14C]hexadecane and the corresponding loss of sulfate that was congruent with the theoretical stoichiometric ratio [11]. Under sulfate-limiting conditions, only 3% of the total alkane present was degraded, demonstrating the dependence of alkane degradation on sulfate reduction [10]. When sulfate concentrations were sufficient, 91% of the alkane present was degraded [10]. The mechanism of initiation via addition to fumarate is not unique to D. alkenivorans AK-01. It has also been observed in the sulfate-reducer Desulfoglaeba alkanexedens [6], Desulfatibacillum aliphaticivorans CV2803 [5], and the denitrifying strain HxN1, a member of the Betaproteobacteria [14]. This reaction involving fumarate represents an alternative mechanism of alkane activation compared to the more commonly known hydroxylation reaction by monooxygenases in aerobic environments [11].
Ecology
From an evolutionary perspective, D. alkenivorans AK-01 is well adapted to occupy a variety of niches. The discovery of genes related to motility and oxygen detoxification suggests AK-01 may be able to adapt to a greater variety of environmental conditions than previously thought [3]. Furthermore, AK-01 is capable of exhibiting three distinct modes of metabolism, giving it a distinct advantage over more specialized organisms [3]. The microbial interactions of AK-01 in situ have yet to be analyzed. There are no known diseases associated with D. alkenivorans AK-01 [13].
Future Applications
Organisms capable of coupling alkane degradation with sulfate reduction play a key role in sites contaminated with hydrocarbons, as these sites typically become anoxic over time due to aerobic respiration and decomposition [4]. Additionally, sulfate is relatively abundant in marine waters compared to nitrate and iron, which typically exist at low concentrations [9]. In one study, analysis of sediment polluted with petroleum after 502 days showed strains most closely related to AK-01 were the most abundant [9], further emphasizing possible bio-remedial applications of D. alkenivorans AK-01 in the future. It has been experimentally shown that AK-01 can interact with methanogens to degrade alkanes syntrophically. This mechanism could be applied within the oil industry, as the current techniques used to retrieve fossil fuels are only able to extract 40% of the available resource [8]. The residual oil could be used to harvest methane gas through syntrophic degradation as an alternative energy source [8]. Current experiments expect a yield of 3 mmol methane gas per gram of residual oil. Taking into account the number of U.S. oil reservoirs, syntrophic alkane degradation could provide 17% of the natural gas consumed in the United States [8].
References
1. Callaghan, A., Gieg, L., Kropp, K., Suflita, J. and Young, L. “Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium.” Appl Environ Microbiol, 2006, DOI: 10.1128/AEM02896-05
2. Callaghan, A., Wawrik, B., Ní Chadhain, S., Young, L. and Zylstra, G. “Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes.” Biochem Biophys Res Commun, 2008, DOI: 10.1016/j.bbrc.2007.11.094
3. Callaghan, A., Morris, B., Pereira, I., McInerney, M., Austin, R., Groves, J., Kukor, J., Suflita, J., Young, L., Zylstra, G. and Wawrik, B. “The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation.” Environ Microbiol, 2012, DOI: 10.1111/j.1462-2920.2011.02516.x
4. Canfield, D., Jorgensen, B., Fossing, H., Glud, R., Gundersen, J., Ramsing, N., Thamdrup, B., Hansen, J., Nielsen, L. and Hall, P. “Pathways of organic carbon oxidation in three continental margin sediments.” Mar Geol, 1993, DOI: 10.1016/0025-3227(93)90147-N
5. Cravo-Laureau, C., Matheron, R., Cayol, J., Joulian, C. and Hirschler-Réa, A. “Desulfatibacillum aliphaticivorans gen. nov., sp. nov., an n-alkane- and n-alkene-degrading, sulfate-reducing bacterium.” Int J Syst Evol Microbiol, 2004, DOI: 10.1099/ijs.0.02717-0
6. Davidova, I., Duncan, K., Choi, O. and Suflita, J. “Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane-degrading, sulfate-reducing bacterium.” Int J Syst Evol Microbiol, 2006, DOI: 10.1099/ijs.0.64398-0
7. Ferry, J., Smith, P. and Wolfe, R. “Methanospirillum, new genus of methanogenic bacteria, and characterization of Methanospirillum hungatei sp. nov.” Int J Syst Evol Microbiol, 1974, Retrieved from http://ijs.sgmjournals.org/content/24/4/465
8. Gieg, L., Duncan, K. and Suflita, J. “Bioenergy Production via Microbial Conversion of Residual Oil to Natural Gas.” Appl Environ Microbiol, 2008, DOI: 10.1128/AEM.00119-08
9. Miralles, G., Grossi, V., Acquaviva, M., Duran, R., Bertrand, J. and Cuny, P. “Alkane biodegradation and dynamics of phylogenetic subgroups of sulfate-reducing bacteria in an anoxic costal marine sediment artificially contaminated with oil.” Chemosphere, 2007, DOI: 10.1016/j.chemosphere.2007.01.033
10. So, C. and Young, L. “Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes.” Appl Environ Microbiol, 1999, Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC91444/
11. So, C. and Young, L. “Initial reactions in anaerobic alkane degradation by a sulfate reducer, strain AK-01.” Appl Environ Microbiol, 1999, Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC91754/
12. So, C., Phelps, C. and Young, L. “Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, strain Hxd3.” Appl Environ Microbiol, 2003, DOI: 10.1128/AEM.69.7.3892-3900.2003
13. U.S. Department of Energy Joint Genome Institute. “Desulfatibacillum alkenivorans AK-01.” 2010, Retrieved from http://genome.jgi.doe.gov/desa1/desa1.info.html
14. Wilkes, H., Rabus, R. and Fischer, T. “Anaerobic degradation of n-hexane in a denitrifying bacterium: Further degradation of the initial intermediate (1-methylpentyl)succinate via C-skeleton rearrangement.” Arch Microbiol, 2002, DOI: 10.1007/s00203-001-0381-3
15. Davidova, I., Duncan, K., Choi, O. and Suflita, J. "Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane-degrading, sulfate-reducing bacterium." Int J Syst Evol Microbiol, 2005, DOI: 10.1099/ijs.0.64398-0